Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems
- PMID: 27095377
- PMCID: PMC4837390
- DOI: 10.1038/srep24645
Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems
Abstract
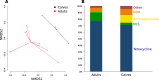
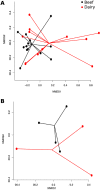
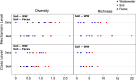
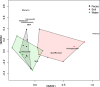
It has been proposed that livestock production effluents such as wastewater, airborne dust and manure increase the density of antimicrobial resistant bacteria and genes in the environment. The public health risk posed by this proposed outcome has been difficult to quantify using traditional microbiological approaches. We utilized shotgun metagenomics to provide a first description of the resistome of North American dairy and beef production effluents, and identify factors that significantly impact this resistome. We identified 34 mechanisms of antimicrobial drug resistance within 34 soil, manure and wastewater samples from feedlot, ranch and dairy operations. The majority of resistance-associated sequences found in all samples belonged to tetracycline resistance mechanisms. We found that the ranch samples contained significantly fewer resistance mechanisms than dairy and feedlot samples, and that the resistome of dairy operations differed significantly from that of feedlots. The resistome in soil, manure and wastewater differed, suggesting that management of these effluents should be tailored appropriately. By providing a baseline of the cattle production waste resistome, this study represents a solid foundation for future efforts to characterize and quantify the public health risk posed by livestock effluents.
Figures


References
-
- Sura S. et al. Transport of three veterinary antimicrobials from feedlot pens via simulated rainfall runoff. Sci. Total Environ. 521, 191–199 (2015). - PubMed
-
- McNab Walt W., Singleton M. J., Moran J. E. & Esser B. K. Assessing the Impact of Animal Waste Lagoon Seepage on the Geochemistry of an Underlying Shallow Aquifer. Environ. Sci. Technol. 41, 753–758 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

