Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells
- PMID: 27096368
- PMCID: PMC4860026
- DOI: 10.1038/nature17950
Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells
Erratum in
-
Corrigendum: Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells.Nature. 2016 Nov 3;539(7627):124. doi: 10.1038/nature19837. Epub 2016 Sep 28. Nature. 2016. PMID: 27680703 No abstract available.
Retraction in
-
Retraction Note: Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells.Nature. 2022 Sep;609(7927):640. doi: 10.1038/s41586-022-05201-2. Nature. 2022. PMID: 36045298 Free PMC article. No abstract available.
Abstract
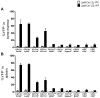
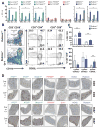
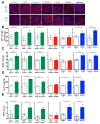
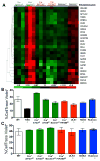
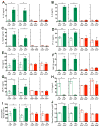
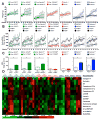
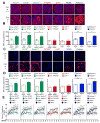
Defects in clearance of dying cells have been proposed to underlie the pathogenesis of systemic lupus erythematosus (SLE). Mice lacking molecules associated with dying cell clearance develop SLE-like disease, and phagocytes from patients with SLE often display defective clearance and increased inflammatory cytokine production when exposed to dying cells in vitro. Previously, we and others described a form of noncanonical autophagy known as LC3-associated phagocytosis (LAP), in which phagosomes containing engulfed particles, including dying cells, recruit elements of the autophagy pathway to facilitate maturation of phagosomes and digestion of their contents. Genome-wide association studies have identified polymorphisms in the Atg5 (ref. 8) and possibly Atg7 (ref. 9) genes, involved in both canonical autophagy and LAP, as markers of a predisposition for SLE. Here we describe the consequences of defective LAP in vivo. Mice lacking any of several components of the LAP pathway show increased serum levels of inflammatory cytokines and autoantibodies, glomerular immune complex deposition, and evidence of kidney damage. When dying cells are injected into LAP-deficient mice, they are engulfed but not efficiently degraded and trigger acute elevation of pro-inflammatory cytokines but not anti-inflammatory interleukin (IL)-10. Repeated injection of dying cells into LAP-deficient, but not LAP-sufficient, mice accelerated the development of SLE-like disease, including increased serum levels of autoantibodies. By contrast, mice deficient in genes required for canonical autophagy but not LAP do not display defective dying cell clearance, inflammatory cytokine production, or SLE-like disease, and, like wild-type mice, produce IL-10 in response to dying cells. Therefore, defects in LAP, rather than canonical autophagy, can cause SLE-like phenomena, and may contribute to the pathogenesis of SLE.
Conflict of interest statement
The authors declare no competing financial interests.
Figures







Comment in
-
Systemic lupus erythematosus: Defective noncanonical autophagy in SLE-like disease.Nat Rev Nephrol. 2016 Jul;12(7):375. doi: 10.1038/nrneph.2016.67. Epub 2016 May 3. Nat Rev Nephrol. 2016. PMID: 27140848 No abstract available.
-
Systemic lupus erythematosus: Defective noncanonical autophagy in SLE-like disease.Nat Rev Rheumatol. 2016 Jun;12(6):311. doi: 10.1038/nrrheum.2016.73. Epub 2016 May 6. Nat Rev Rheumatol. 2016. PMID: 27150664 No abstract available.
-
Non-canonical autophagy LAPs lupus.Cell Death Differ. 2016 Aug;23(8):1267-8. doi: 10.1038/cdd.2016.55. Epub 2016 Jun 10. Cell Death Differ. 2016. PMID: 27285105 Free PMC article. No abstract available.
-
LAP: the protector against autoimmunity.Cell Res. 2016 Aug;26(8):865-6. doi: 10.1038/cr.2016.70. Epub 2016 Jun 14. Cell Res. 2016. PMID: 27297234 Free PMC article.
-
LAPping up dead cells to prevent lupus nephritis: a novel role for noncanonical autophagy in autoimmunity.Kidney Int. 2016 Aug;90(2):238-239. doi: 10.1016/j.kint.2016.06.001. Kidney Int. 2016. PMID: 27418084
-
Protection against lupus-like inflammatory disease is in the LAP of non-canonical autophagy.Ann Transl Med. 2016 Dec;4(24):513. doi: 10.21037/atm.2016.12.30. Ann Transl Med. 2016. PMID: 28149875 Free PMC article. No abstract available.
References
-
- Rodriguez-Manzanet R, et al. T and B cell hyperactivity and autoimmunity associated with niche-specific defects in apoptotic body clearance in TIM-4-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8706–8711. doi: 10.1073/pnas.0910359107. - DOI - PMC - PubMed
-
- Martinez J, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17396–17401. doi: 10.1073/pnas.1113421108. - DOI - PMC - PubMed
-
- Martinez JM, RK, Lu Q, Cunha LD, Pelletier S, Gingras S, Orchard R, Guan JL, Tan H, Peng J, Kanneganti TD, Virgin HW, Green DR. Molecular characterization of LC3-assMolecular characterization of LC3-associated phagocytosis (LAP) reveals distinct roles for Rubicon, NOX2, and autophagy proteins. Nature cell biology. 2015 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

