Learning Biomarker Models for Progression Estimation of Alzheimer's Disease
- PMID: 27096739
- PMCID: PMC4838309
- DOI: 10.1371/journal.pone.0153040
Learning Biomarker Models for Progression Estimation of Alzheimer's Disease
Abstract
Being able to estimate a patient's progress in the course of Alzheimer's disease and predicting future progression based on a number of observed biomarker values is of great interest for patients, clinicians and researchers alike. In this work, an approach for disease progress estimation is presented. Based on a set of subjects that convert to a more severe disease stage during the study, models that describe typical trajectories of biomarker values in the course of disease are learned using quantile regression. A novel probabilistic method is then derived to estimate the current disease progress as well as the rate of progression of an individual by fitting acquired biomarkers to the models. A particular strength of the method is its ability to naturally handle missing data. This means, it is applicable even if individual biomarker measurements are missing for a subject without requiring a retraining of the model. The functionality of the presented method is demonstrated using synthetic and--employing cognitive scores and image-based biomarkers--real data from the ADNI study. Further, three possible applications for progress estimation are demonstrated to underline the versatility of the approach: classification, construction of a spatio-temporal disease progression atlas and prediction of future disease progression.
Conflict of interest statement
Figures
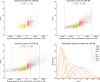









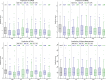
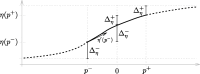
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

