Evidence of adaptation, niche separation and microevolution within the genus Polaromonas on Arctic and Antarctic glacial surfaces
- PMID: 27097637
- PMCID: PMC4921121
- DOI: 10.1007/s00792-016-0831-0
Evidence of adaptation, niche separation and microevolution within the genus Polaromonas on Arctic and Antarctic glacial surfaces
Abstract
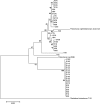
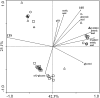
Polaromonas is one of the most abundant genera found on glacier surfaces, yet its ecology remains poorly described. Investigations made to date point towards a uniform distribution of Polaromonas phylotypes across the globe. We compared 43 Polaromonas isolates obtained from surfaces of Arctic and Antarctic glaciers to address this issue. 16S rRNA gene sequences, intergenic transcribed spacers (ITS) and metabolic fingerprinting showed great differences between hemispheres but also between neighboring glaciers. Phylogenetic distance between Arctic and Antarctic isolates indicated separate species. The Arctic group clustered similarly, when constructing dendrograms based on 16S rRNA gene and ITS sequences, as well as metabolic traits. The Antarctic strains, although almost identical considering 16S rRNA genes, diverged into 2 groups based on the ITS sequences and metabolic traits, suggesting recent niche separation. Certain phenotypic traits pointed towards cell adaptation to specific conditions on a particular glacier, like varying pH levels. Collected data suggest, that seeding of glacial surfaces with Polaromonas cells transported by various means, is of greater efficiency on local than global scales. Selection mechanisms present of glacial surfaces reduce the deposited Polaromonas diversity, causing subsequent adaptation to prevailing environmental conditions. Furthermore, interactions with other supraglacial microbiota, like algae cells may drive postselectional niche separation and microevolution within the Polaromonas genus.
Keywords: 16S rRNA gene; Bacteria; Biogeography; Glacier; ITS; Polaromonas.
Figures




Similar articles
-
Plasmids of Psychrotolerant Polaromonas spp. Isolated From Arctic and Antarctic Glaciers - Diversity and Role in Adaptation to Polar Environments.Front Microbiol. 2018 Jun 18;9:1285. doi: 10.3389/fmicb.2018.01285. eCollection 2018. Front Microbiol. 2018. PMID: 29967598 Free PMC article.
-
Polaromonas glacialis sp. nov. and Polaromonas cryoconiti sp. nov., isolated from alpine glacier cryoconite.Int J Syst Evol Microbiol. 2012 Nov;62(Pt 11):2662-2668. doi: 10.1099/ijs.0.037556-0. Epub 2011 Dec 23. Int J Syst Evol Microbiol. 2012. PMID: 22199222
-
The dynamic bacterial communities of a melting High Arctic glacier snowpack.ISME J. 2013 Sep;7(9):1814-26. doi: 10.1038/ismej.2013.51. Epub 2013 Apr 4. ISME J. 2013. PMID: 23552623 Free PMC article.
-
Global distribution of Polaromonas phylotypes--evidence for a highly successful dispersal capacity.PLoS One. 2011;6(8):e23742. doi: 10.1371/journal.pone.0023742. Epub 2011 Aug 29. PLoS One. 2011. PMID: 21897856 Free PMC article.
-
Psychrophilic yeasts from worldwide glacial habitats: diversity, adaptation strategies and biotechnological potential.FEMS Microbiol Ecol. 2012 Nov;82(2):217-41. doi: 10.1111/j.1574-6941.2012.01348.x. Epub 2012 Mar 27. FEMS Microbiol Ecol. 2012. PMID: 22385361 Review.
Cited by
-
Bacterial community structure in a sympagic habitat expanding with global warming: brackish ice brine at 85-90 °N.ISME J. 2019 Feb;13(2):316-333. doi: 10.1038/s41396-018-0268-9. Epub 2018 Sep 18. ISME J. 2019. PMID: 30228379 Free PMC article.
-
Isolation of Acanthamoeba Species and Bacterial Symbiont Variability in Puna Salt Plains, Argentina.Environ Microbiol Rep. 2025 Feb;17(1):e70059. doi: 10.1111/1758-2229.70059. Environ Microbiol Rep. 2025. PMID: 39810455 Free PMC article.
-
H2 Metabolism revealed by metagenomic analysis of subglacial sediment from East Antarctica.J Microbiol. 2019 Dec;57(12):1095-1104. doi: 10.1007/s12275-019-9366-2. Epub 2019 Nov 22. J Microbiol. 2019. PMID: 31758395
-
Bacterioplankton seasonality in deep high-mountain lakes.Front Microbiol. 2022 Sep 14;13:935378. doi: 10.3389/fmicb.2022.935378. eCollection 2022. Front Microbiol. 2022. PMID: 36187988 Free PMC article.
-
Bacteria of the order Burkholderiales are original environmental hosts of type II trimethoprim resistance genes (dfrB).ISME J. 2024 Jan 8;18(1):wrae243. doi: 10.1093/ismejo/wrae243. ISME J. 2024. PMID: 39658215 Free PMC article.
References
-
- Anesio AM, Hodson AJ, Fritz A, Psenner R, Sattler B. High microbial activity on glaciers: importance to the global carbon cycle. Glob Change Biol. 2009;15:955–960. doi: 10.1111/j.1365-2486.2008.01758.x. - DOI
-
- Brown MV, Fuhrman JA. Marine bacterial microdiversity as revealed by internal transcribed spacer analysis. Aquat Microb Ecol. 2005;41:15–23. doi: 10.3354/ame041015. - DOI
-
- Brown MV, Schwalbach MS, Hewson I, Fuhrman JA. Coupling 16S-ITS rRNA gene clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ Microbiol. 2005;7(9):1466–1479. doi: 10.1111/j.1462-2920.2005.00835.x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

