Perturbed rhythmic activation of signaling pathways in mice deficient for Sterol Carrier Protein 2-dependent diurnal lipid transport and metabolism
- PMID: 27097688
- PMCID: PMC4838911
- DOI: 10.1038/srep24631
Perturbed rhythmic activation of signaling pathways in mice deficient for Sterol Carrier Protein 2-dependent diurnal lipid transport and metabolism
Abstract
Through evolution, most of the living species have acquired a time keeping system to anticipate daily changes caused by the rotation of the Earth. In all of the systems this pacemaker is based on a molecular transcriptional/translational negative feedback loop able to generate rhythmic gene expression with a period close to 24 hours. Recent evidences suggest that post-transcriptional regulations activated mostly by systemic cues play a fundamental role in the process, fine tuning the time keeping system and linking it to animal physiology. Among these signals, we consider the role of lipid transport and metabolism regulated by SCP2. Mice harboring a deletion of the Scp2 locus present a modulated diurnal accumulation of lipids in the liver and a perturbed activation of several signaling pathways including PPARα, SREBP, LRH-1, TORC1 and its upstream regulators. This defect in signaling pathways activation feedbacks upon the clock by lengthening the circadian period of animals through post-translational regulation of core clock regulators, showing that rhythmic lipid transport is a major player in the establishment of rhythmic mRNA and protein expression landscape.
Conflict of interest statement
All authors are employees of Nestlé Institute of Health Sciences SA.
Figures
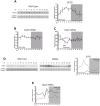
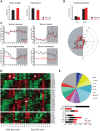
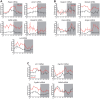
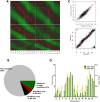
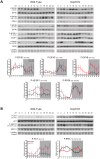
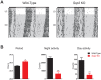
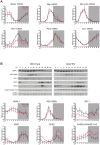
Similar articles
-
Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms.Proc Natl Acad Sci U S A. 2021 Jan 19;118(3):e2015803118. doi: 10.1073/pnas.2015803118. Proc Natl Acad Sci U S A. 2021. PMID: 33452134 Free PMC article.
-
Rewiring of liver diurnal transcriptome rhythms by triiodothyronine (T3) supplementation.Elife. 2022 Jul 27;11:e79405. doi: 10.7554/eLife.79405. Elife. 2022. PMID: 35894384 Free PMC article.
-
REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis.PLoS Biol. 2009 Sep;7(9):e1000181. doi: 10.1371/journal.pbio.1000181. Epub 2009 Sep 1. PLoS Biol. 2009. PMID: 19721697 Free PMC article.
-
Post-transcriptional modulators and mediators of the circadian clock.Chronobiol Int. 2021 Sep;38(9):1244-1261. doi: 10.1080/07420528.2021.1928159. Epub 2021 May 31. Chronobiol Int. 2021. PMID: 34056966 Free PMC article. Review.
-
Post-transcriptional control of circadian rhythms.J Cell Sci. 2011 Feb 1;124(Pt 3):311-20. doi: 10.1242/jcs.065771. J Cell Sci. 2011. PMID: 21242310 Free PMC article. Review.
Cited by
-
Circadian- and UPR-dependent control of CPEB4 mediates a translational response to counteract hepatic steatosis under ER stress.Nat Cell Biol. 2017 Feb;19(2):94-105. doi: 10.1038/ncb3461. Epub 2017 Jan 16. Nat Cell Biol. 2017. PMID: 28092655
-
Transcriptomics-driven metabolic pathway analysis reveals similar alterations in lipid metabolism in mouse MASH model and human.Commun Med (Lond). 2024 Mar 5;4(1):39. doi: 10.1038/s43856-024-00465-3. Commun Med (Lond). 2024. PMID: 38443644 Free PMC article.
-
Reproducibility across single-cell RNA-seq protocols for spatial ordering analysis.PLoS One. 2020 Sep 28;15(9):e0239711. doi: 10.1371/journal.pone.0239711. eCollection 2020. PLoS One. 2020. PMID: 32986734 Free PMC article.
-
Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms.Proc Natl Acad Sci U S A. 2021 Jan 19;118(3):e2015803118. doi: 10.1073/pnas.2015803118. Proc Natl Acad Sci U S A. 2021. PMID: 33452134 Free PMC article.
-
Transcriptional programming of lipid and amino acid metabolism by the skeletal muscle circadian clock.PLoS Biol. 2018 Aug 10;16(8):e2005886. doi: 10.1371/journal.pbio.2005886. eCollection 2018 Aug. PLoS Biol. 2018. PMID: 30096135 Free PMC article.
References
-
- Asher G. & Schibler U. Crosstalk between Components of Circadian and Metabolic Cycles in Mammals. Cell Metab 13, 125–137, doi: 10.1016/j.cmet.2011.01.006 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

