Real-time quantification of protein expression at the single-cell level via dynamic protein synthesis translocation reporters
- PMID: 27098003
- PMCID: PMC4844680
- DOI: 10.1038/ncomms11304
Real-time quantification of protein expression at the single-cell level via dynamic protein synthesis translocation reporters
Abstract
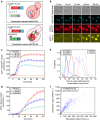
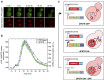
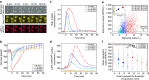
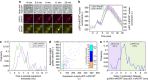
Protein expression is a dynamic process, which can be rapidly induced by extracellular signals. It is widely appreciated that single cells can display large variations in the level of gene induction. However, the variability in the dynamics of this process in individual cells is difficult to quantify using standard fluorescent protein (FP) expression assays, due to the slow maturation of their fluorophore. Here we have developed expression reporters that accurately measure both the levels and dynamics of protein synthesis in live single cells with a temporal resolution under a minute. Our system relies on the quantification of the translocation of a constitutively expressed FP into the nucleus. As a proof of concept, we used these reporters to measure the transient protein synthesis arising from two promoters responding to the yeast hyper osmolarity glycerol mitogen-activated protein kinase pathway (pSTL1 and pGPD1). They display distinct expression dynamics giving rise to strikingly different instantaneous expression noise.
Figures
References
-
- de Nadal E., Ammerer G. & Posas F. Controlling gene expression in response to stress. Nat. Rev. Genet. 12, 833–845 (2011) . - PubMed
-
- Weake V. M. & Workman J. L. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 11, 426–437 (2010) . - PubMed
-
- Kadonaga J. T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116, 247–257 (2004) . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

