Towards Development of Improved Serodiagnostics for Tularemia by Use of Francisella tularensis Proteome Microarrays
- PMID: 27098957
- PMCID: PMC4922089
- DOI: 10.1128/JCM.02784-15
Towards Development of Improved Serodiagnostics for Tularemia by Use of Francisella tularensis Proteome Microarrays
Abstract
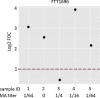
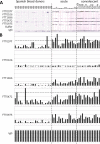
Tularemia in humans is caused mainly by two subspecies of the Gram-negative facultative anaerobe Francisella tularensis: F. tularensis subsp. tularensis (type A) and F. tularensis subsp. holarctica (type B). The current serological test for tularemia is based on agglutination of whole organisms, and the reactive antigens are not well understood. Previously, we profiled the antibody responses in type A and B tularemia cases in the United States using a proteome microarray of 1,741 different proteins derived from the type A strain Schu S4. Fifteen dominant antigens able to detect antibodies to both types of infection were identified, although these were not validated in a different immunoassay format. Since type A and B subspecies are closely related, we hypothesized that Schu S4 antigens would also have utility for diagnosing type B tularemia caused by strains from other geographic locations. To test this, we probed the Schu S4 array with sera from 241 type B tularemia cases in Spain. Despite there being no type A strains in Spain, we confirmed the responses against some of the same potential serodiagnostic antigens reported previously, as well as determined the responses against additional potential serodiagnostic antigens. Five potential serodiagnostic antigens were evaluated on immunostrips, and two of these (FTT1696/GroEL and FTT0975/conserved hypothetical protein) discriminated between the Spanish tularemia cases and healthy controls. We conclude that antigens from the type A strain Schu S4 are suitable for detection of antibodies from patients with type B F. tularensis infections and that these can be used for the diagnosis of tularemia in a deployable format, such as the immunostrip.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Characterization of Schu S4 aro mutants as live attenuated tularemia vaccine candidates.Virulence. 2020 Dec;11(1):283-294. doi: 10.1080/21505594.2020.1746557. Virulence. 2020. PMID: 32241221 Free PMC article.
-
Long-Term Survival of Virulent Tularemia Pathogens outside a Host in Conditions That Mimic Natural Aquatic Environments.Appl Environ Microbiol. 2021 Feb 26;87(6):e02713-20. doi: 10.1128/AEM.02713-20. Print 2021 Feb 26. Appl Environ Microbiol. 2021. PMID: 33397692 Free PMC article.
-
Evaluation of In-House and Commercial Serological Tests for Diagnosis of Human Tularemia.J Clin Microbiol. 2017 Dec 26;56(1):e01440-17. doi: 10.1128/JCM.01440-17. Print 2018 Jan. J Clin Microbiol. 2017. PMID: 29118164 Free PMC article.
-
Francisella tularensis: taxonomy, genetics, and Immunopathogenesis of a potential agent of biowarfare.Annu Rev Microbiol. 2006;60:167-85. doi: 10.1146/annurev.micro.60.080805.142126. Annu Rev Microbiol. 2006. PMID: 16704343 Free PMC article. Review.
-
[Diagnostic tests: Tularemia].Nihon Rinsho. 2005 Jul;63 Suppl 7:253-5. Nihon Rinsho. 2005. PMID: 16111241 Review. Japanese. No abstract available.
Cited by
-
Shotgun Immunoproteomic Approach for the Discovery of Linear B-Cell Epitopes in Biothreat Agents Francisella tularensis and Burkholderia pseudomallei.Front Immunol. 2021 Sep 29;12:716676. doi: 10.3389/fimmu.2021.716676. eCollection 2021. Front Immunol. 2021. PMID: 34659206 Free PMC article.
-
Transient Cannabinoid Receptor 2 Blockade during Immunization Heightens Intensity and Breadth of Antigen-specific Antibody Responses in Young and Aged mice.Sci Rep. 2017 Feb 17;7:42584. doi: 10.1038/srep42584. Sci Rep. 2017. PMID: 28209996 Free PMC article.
-
Challenges of Francisella classification exemplified by an atypical clinical isolate.Diagn Microbiol Infect Dis. 2018 Apr;90(4):241-247. doi: 10.1016/j.diagmicrobio.2017.11.023. Epub 2017 Dec 6. Diagn Microbiol Infect Dis. 2018. PMID: 29329757 Free PMC article.
-
Francisella tularensis, Tularemia and Serological Diagnosis.Front Cell Infect Microbiol. 2020 Oct 26;10:512090. doi: 10.3389/fcimb.2020.512090. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33194778 Free PMC article. Review.
-
Development of ELISAs for diagnosis of acute typhoid fever in Nigerian children.PLoS Negl Trop Dis. 2017 Jun 22;11(6):e0005679. doi: 10.1371/journal.pntd.0005679. eCollection 2017 Jun. PLoS Negl Trop Dis. 2017. PMID: 28640809 Free PMC article.
References
-
- Centro Nacional de Epidemiología Instituto de Salud Carlos III. 1997. Brote de tularemia en Castilla y León. Bol Epidemiol Sem 5:249–252.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

