UBIQUITIN-SPECIFIC PROTEASE14 Interacts with ULTRAVIOLET-B INSENSITIVE4 to Regulate Endoreduplication and Cell and Organ Growth in Arabidopsis
- PMID: 27099260
- PMCID: PMC4904675
- DOI: 10.1105/tpc.16.00007
UBIQUITIN-SPECIFIC PROTEASE14 Interacts with ULTRAVIOLET-B INSENSITIVE4 to Regulate Endoreduplication and Cell and Organ Growth in Arabidopsis
Abstract
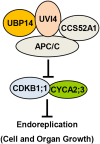
Organ growth is determined by a coordinated combination of cell proliferation and cell growth and differentiation. Endoreduplication is often coupled with cell growth and differentiation, but the genetic and molecular mechanisms that link endoreduplication with cell and organ growth are largely unknown. Here, we describe UBIQUITIN-SPECIFIC PROTEASE14 (UBP14), encoded by the DA3 gene, which functions as a negative regulator of endoreduplication. The Arabidopsis thaliana da3-1 mutant shows large cotyledons, leaves, and flowers with higher ploidy levels. UBP14 acts along with UV-B-INSENSITIVE4 (UVI4), an inhibitor of the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase, to repress endoreduplication. Also, UBP14 functions antagonistically with CELL CYCLE SWITCH52 A1 (CCS52A1), an activator of APC/C, to regulate endoreduplication. UBP14 physically associates with UVI4 both in vitro and in vivo but does not directly interact with CCS52A1. Further results reveal that UBP14 influences the stability of cyclin A2;3 (CYCA2;3) and cyclin-dependent kinase B1;1 (CDKB1;1), two downstream components of the APC/C Thus, our findings show how endoreduplication is linked with cell and organ growth by revealing important genetic and molecular functions for the ubiquitin-specific protease UBP14 and for the key cell cycle regulators UVI4, CCS52A1, CYCA2;3, and CDKB1;1.
© 2016 American Society of Plant Biologists. All rights reserved.
Figures

Similar articles
-
The UBP14-CDKB1;1-CDKG2 cascade controls endoreduplication and cell growth in Arabidopsis.Plant Cell. 2022 Mar 29;34(4):1308-1325. doi: 10.1093/plcell/koac002. Plant Cell. 2022. PMID: 34999895 Free PMC article.
-
An SNW/SKI-INTERACTING PROTEIN influences endoreduplication and cell growth in Arabidopsis.Plant Physiol. 2022 Nov 28;190(4):2217-2228. doi: 10.1093/plphys/kiac415. Plant Physiol. 2022. PMID: 36063458 Free PMC article.
-
Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome.Plant Cell. 2011 Dec;23(12):4394-410. doi: 10.1105/tpc.111.091793. Epub 2011 Dec 13. Plant Cell. 2011. PMID: 22167059 Free PMC article.
-
Insights into phosphorylation-dependent mechanisms regulating USP1 protein stability during the cell cycle.Cell Cycle. 2011 Dec 1;10(23):4009-16. doi: 10.4161/cc.10.23.18501. Epub 2011 Dec 1. Cell Cycle. 2011. PMID: 22101265 Free PMC article. Review.
-
Re-examination of the role of endoreduplication on cell-size control in leaves.J Plant Res. 2019 Sep;132(5):571-580. doi: 10.1007/s10265-019-01125-7. Epub 2019 Jul 18. J Plant Res. 2019. PMID: 31321606 Free PMC article. Review.
Cited by
-
Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis.Nat Commun. 2020 Apr 15;11(1):1846. doi: 10.1038/s41467-020-15603-3. Nat Commun. 2020. PMID: 32296056 Free PMC article.
-
Auxin apical dominance governed by the OsAsp1-OsTIF1 complex determines distinctive rice caryopses development on different branches.PLoS Genet. 2020 Oct 27;16(10):e1009157. doi: 10.1371/journal.pgen.1009157. eCollection 2020 Oct. PLoS Genet. 2020. PMID: 33108367 Free PMC article.
-
The UBP14-CDKB1;1-CDKG2 cascade controls endoreduplication and cell growth in Arabidopsis.Plant Cell. 2022 Mar 29;34(4):1308-1325. doi: 10.1093/plcell/koac002. Plant Cell. 2022. PMID: 34999895 Free PMC article.
-
Plant Deubiquitinases and Their Role in the Control of Gene Expression Through Modification of Histones.Front Plant Sci. 2018 Jan 17;8:2274. doi: 10.3389/fpls.2017.02274. eCollection 2017. Front Plant Sci. 2018. PMID: 29387079 Free PMC article. Review.
-
UBIQUITIN-SPECIFIC PROTEASES function in plant development and stress responses.Plant Mol Biol. 2017 Aug;94(6):565-576. doi: 10.1007/s11103-017-0633-5. Epub 2017 Jul 10. Plant Mol Biol. 2017. PMID: 28695315 Review.
References
-
- Baloban M., Vanstraelen M., Tarayre S., Reuzeau C., Cultrone A., Mergaert P., Kondorosi E. (2013). Complementary and dose-dependent action of AtCCS52A isoforms in endoreduplication and plant size control. New Phytol. 198: 1049–1059. - PubMed
-
- Boudolf V., Vlieghe K., Beemster G.T., Magyar Z., Torres Acosta J.A., Maes S., Van Der Schueren E., Inzé D., De Veylder L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692. - PMC - PubMed
-
- Breuer C., Braidwood L., Sugimoto K. (2014). Endocycling in the path of plant development. Curr. Opin. Plant Biol. 17: 78–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

