Noncanonical DNA-binding mode of repressor and its disassembly by antirepressor
- PMID: 27099293
- PMCID: PMC4983836
- DOI: 10.1073/pnas.1602618113
Noncanonical DNA-binding mode of repressor and its disassembly by antirepressor
Abstract
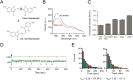
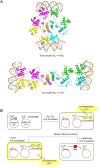
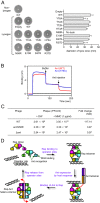
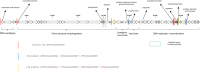
DNA-binding repressors are involved in transcriptional repression in many organisms. Disabling a repressor is a crucial step in activating expression of desired genes. Thus, several mechanisms have been identified for the removal of a stably bound repressor (Rep) from the operator. Here, we describe an uncharacterized mechanism of noncanonical DNA binding and induction by a Rep from the temperate Salmonella phage SPC32H; this mechanism was revealed using the crystal structures of homotetrameric Rep (92-198) and a hetero-octameric complex between the Rep and its antirepressor (Ant). The canonical method of inactivating a repressor is through the competitive binding of the antirepressor to the operator-binding site of the repressor; however, these studies revealed several noncanonical features. First, Ant does not compete for the DNA-binding region of Rep. Instead, the tetrameric Ant binds to the C-terminal domains of two asymmetric Rep dimers. Simultaneously, Ant facilitates the binding of the Rep N-terminal domains to Ant, resulting in the release of two Rep dimers from the bound DNA. Second, the dimer pairs of the N-terminal DNA-binding domains originate from different dimers of a Rep tetramer (trans model). This situation is different from that of other canonical Reps, in which two N-terminal DNA-binding domains from the same dimeric unit form a dimer upon DNA binding (cis model). On the basis of these observations, we propose a noncanonical model for the reversible inactivation of a Rep by an Ant.
Keywords: Salmonella; antirepressor; bacteriophage; repressor; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Carboxyl-terminal domain dimer interface mutant 434 repressors have altered dimerization and DNA binding specificities.J Mol Biol. 1998 Nov 13;283(5):931-46. doi: 10.1006/jmbi.1998.2136. J Mol Biol. 1998. PMID: 9799634
-
Binding of the N-terminal domain of the lactococcal bacteriophage TP901-1 CI repressor to its target DNA: a crystallography, small angle scattering, and nuclear magnetic resonance study.Biochemistry. 2013 Oct 1;52(39):6892-904. doi: 10.1021/bi400439y. Epub 2013 Sep 18. Biochemistry. 2013. PMID: 24047404
-
Antirepression system associated with the life cycle switch in the temperate podoviridae phage SPC32H.J Virol. 2013 Nov;87(21):11775-86. doi: 10.1128/JVI.02173-13. Epub 2013 Aug 28. J Virol. 2013. PMID: 23986584 Free PMC article.
-
Four dimers of lambda repressor bound to two suitably spaced pairs of lambda operators form octamers and DNA loops over large distances.Curr Biol. 1999 Feb 11;9(3):151-4. doi: 10.1016/s0960-9822(99)80069-4. Curr Biol. 1999. PMID: 10021390
-
Identification of quaternary structure and functional domains of the CI repressor from bacteriophage TP901-1.J Mol Biol. 2008 Feb 29;376(4):983-96. doi: 10.1016/j.jmb.2007.12.022. Epub 2007 Dec 15. J Mol Biol. 2008. PMID: 18191944
Cited by
-
A dual-function phage regulator controls the response of cohabiting phage elements via regulation of the bacterial SOS response.Cell Rep. 2022 Apr 19;39(3):110723. doi: 10.1016/j.celrep.2022.110723. Cell Rep. 2022. PMID: 35443160 Free PMC article.
-
Isolation, phenotypic characterization and comparative genomic analysis of 2019SD1, a polyvalent enterobacteria phage.Sci Rep. 2021 Nov 12;11(1):22197. doi: 10.1038/s41598-021-01419-8. Sci Rep. 2021. PMID: 34772986 Free PMC article.
-
A Syringe-Based Biosensor to Rapidly Detect Low Levels of Escherichia Coli (ECOR13) in Drinking Water Using Engineered Bacteriophages.Sensors (Basel). 2020 Mar 31;20(7):1953. doi: 10.3390/s20071953. Sensors (Basel). 2020. PMID: 32244369 Free PMC article.
-
The phage gene wmk is a candidate for male killing by a bacterial endosymbiont.PLoS Pathog. 2019 Sep 10;15(9):e1007936. doi: 10.1371/journal.ppat.1007936. eCollection 2019 Sep. PLoS Pathog. 2019. PMID: 31504075 Free PMC article.
-
An archaeal transcription factor EnfR with a novel 'eighth note' fold controls hydrogen production of a hyperthermophilic archaeon Thermococcus onnurineus NA1.Nucleic Acids Res. 2023 Oct 13;51(18):10026-10040. doi: 10.1093/nar/gkad699. Nucleic Acids Res. 2023. PMID: 37650645 Free PMC article.
References
-
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2(1):57–65. - PubMed
-
- Oppenheim AB, Neubauer Z, Calef E. The antirepressor: A new element in the regulation of protein synthesis. Nature. 1970;226(5240):31–32. - PubMed
-
- Luo Y, et al. Crystal structure of LexA: A conformational switch for regulation of self-cleavage. Cell. 2001;106(5):585–594. - PubMed
-
- Wang HC, Ho CH, Hsu KC, Yang JM, Wang AH. DNA mimic proteins: Functions, structures, and bioinformatic analysis. Biochemistry. 2014;53(18):2865–2874. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

