Interferon Alpha Subtype-Specific Suppression of HIV-1 Infection In Vivo
- PMID: 27099312
- PMCID: PMC4907223
- DOI: 10.1128/JVI.00451-16
Interferon Alpha Subtype-Specific Suppression of HIV-1 Infection In Vivo
Abstract
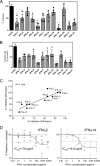
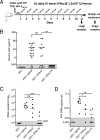
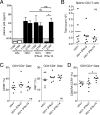
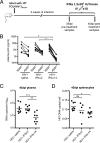
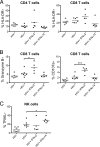
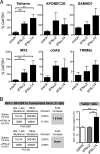
Although all 12 subtypes of human interferon alpha (IFN-α) bind the same receptor, recent results have demonstrated that they elicit unique host responses and display distinct efficacies in the control of different viral infections. The IFN-α2 subtype is currently in HIV-1 clinical trials, but it has not consistently reduced viral loads in HIV-1 patients and is not the most effective subtype against HIV-1 in vitro We now demonstrate in humanized mice that, when delivered at the same high clinical dose, the human IFN-α14 subtype has very potent anti-HIV-1 activity whereas IFN-α2 does not. In both postexposure prophylaxis and treatment of acute infections, IFN-α14, but not IFN-α2, significantly suppressed HIV-1 replication and proviral loads. Furthermore, HIV-1-induced immune hyperactivation, which is a prognosticator of disease progression, was reduced by IFN-α14 but not IFN-α2. Whereas ineffective IFN-α2 therapy was associated with CD8(+) T cell activation, successful IFN-α14 therapy was associated with increased intrinsic and innate immunity, including significantly higher induction of tetherin and MX2, increased APOBEC3G signature mutations in HIV-1 proviral DNA, and higher frequencies of TRAIL(+) NK cells. These results identify IFN-α14 as a potent new therapeutic that operates via mechanisms distinct from those of antiretroviral drugs. The ability of IFN-α14 to reduce both viremia and proviral loads in vivo suggests that it has strong potential as a component of a cure strategy for HIV-1 infections. The broad implication of these results is that the antiviral efficacy of each individual IFN-α subtype should be evaluated against the specific virus being treated.
Importance: The naturally occurring antiviral protein IFN-α2 is used to treat hepatitis viruses but has proven rather ineffective against HIV in comparison to triple therapy with the antiretroviral (ARV) drugs. Although ARVs suppress the replication of HIV, they fail to completely clear infections. Since IFN-α acts by different mechanism than ARVs and has been shown to reduce HIV proviral loads, clinical trials are under way to test whether IFN-α2 combined with ARVs might eradicate HIV-1 infections. IFN-α is actually a family of 12 distinct proteins, and each IFN-α subtype has different efficacies toward different viruses. Here, we use mice that contain a human immune system, so they can be infected with HIV. With this model, we demonstrate that while IFN-α2 is only weakly effective against HIV, IFN-α14 is extremely potent. This discovery identifies IFN-α14 as a more powerful IFN-α subtype for use in combination therapy trials aimed toward an HIV cure.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A, Lee C, Yarden G, Vleck SE, Glenn JS, Nolan GP, Piehler J, Schreiber G, Garcia KC. 2011. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146:621–632. doi:10.1016/j.cell.2011.06.048. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

