Molecular Signatures of Membrane Protein Complexes Underlying Muscular Dystrophy
- PMID: 27099343
- PMCID: PMC5083101
- DOI: 10.1074/mcp.M116.059188
Molecular Signatures of Membrane Protein Complexes Underlying Muscular Dystrophy
Abstract
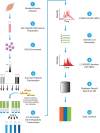
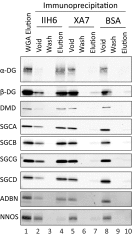
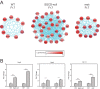
Mutations in genes encoding components of the sarcolemmal dystrophin-glycoprotein complex (DGC) are responsible for a large number of muscular dystrophies. As such, molecular dissection of the DGC is expected to both reveal pathological mechanisms, and provides a biological framework for validating new DGC components. Establishment of the molecular composition of plasma-membrane protein complexes has been hampered by a lack of suitable biochemical approaches. Here we present an analytical workflow based upon the principles of protein correlation profiling that has enabled us to model the molecular composition of the DGC in mouse skeletal muscle. We also report our analysis of protein complexes in mice harboring mutations in DGC components. Bioinformatic analyses suggested that cell-adhesion pathways were under the transcriptional control of NFκB in DGC mutant mice, which is a finding that is supported by previous studies that showed NFκB-regulated pathways underlie the pathophysiology of DGC-related muscular dystrophies. Moreover, the bioinformatic analyses suggested that inflammatory and compensatory mechanisms were activated in skeletal muscle of DGC mutant mice. Additionally, this proteomic study provides a molecular framework to refine our understanding of the DGC, identification of protein biomarkers of neuromuscular disease, and pharmacological interrogation of the DGC in adult skeletal muscle https://www.mda.org/disease/congenital-muscular-dystrophy/research.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Mokri B., and Engel A. G. (1975) Duchenne dystrophy: electron microscopic findings pointing to a basic or early abnormality in the plasma membrane of the muscle fiber. Neurology 25, 1111–1120 - PubMed
-
- Ibraghimov-Beskrovnaya O., Ervasti J. M., Leveille C. J., Slaughter C. A., Sernett S. W., and Campbell K. P. (1992) Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature 355, 696–702 - PubMed
-
- Hohenester E., Tisi D., Talts J. F., and Timpl R. (1999) The crystal structure of a laminin G-like module reveals the molecular basis of alpha-dystroglycan binding to laminins, perlecan, and agrin. Mol. Cell 4, 783–792 - PubMed
-
- Campbell K. P., and Kahl S. D. (1989) Association of dystrophin and an integral membrane glycoprotein. Nature 338, 259–262 - PubMed
-
- Ozawa E., Yoshida M., Suzuki A., Mizuno Y., Hagiwara Y., and Noguchi S. (1995) Dystrophin-associated proteins in muscular dystrophy. Human Mol. Genetics 4, 1711–1716 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

