The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits
- PMID: 27099397
- PMCID: PMC4878195
- DOI: 10.1194/jlr.R066944
The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits
Abstract
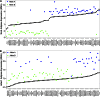
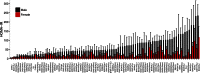
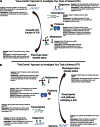
The Hybrid Mouse Diversity Panel (HMDP) is a collection of approximately 100 well-characterized inbred strains of mice that can be used to analyze the genetic and environmental factors underlying complex traits. While not nearly as powerful for mapping genetic loci contributing to the traits as human genome-wide association studies, it has some important advantages. First, environmental factors can be controlled. Second, relevant tissues are accessible for global molecular phenotyping. Finally, because inbred strains are renewable, results from separate studies can be integrated. Thus far, the HMDP has been studied for traits relevant to obesity, diabetes, atherosclerosis, osteoporosis, heart failure, immune regulation, fatty liver disease, and host-gut microbiota interactions. High-throughput technologies have been used to examine the genomes, epigenomes, transcriptomes, proteomes, metabolomes, and microbiomes of the mice under various environmental conditions. All of the published data are available and can be readily used to formulate hypotheses about genes, pathways and interactions.
Keywords: aherosclerosis; gene expression; gene mapping; gene-by-diet interaction; heart failure; insulin resistance; microbiota; obesity; osteoporosis.
Copyright © 2016 by the American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

