Novel, In-House, SYBR Green Based One-Step rRT-PCR: Rapid and Accurate Diagnosis of Crimean-Congo Hemorrhagic Fever Virus in Suspected Patients From Iran
- PMID: 27099688
- PMCID: PMC4834134
- DOI: 10.5812/jjm.29246
Novel, In-House, SYBR Green Based One-Step rRT-PCR: Rapid and Accurate Diagnosis of Crimean-Congo Hemorrhagic Fever Virus in Suspected Patients From Iran
Abstract
Background: The Crimean-Congo hemorrhagic fever (CCHF) virus causes severe disease in humans, with a high mortality rate. Since, there is no approved vaccine or specific treatment for CCHF, an early and accurate diagnosis, as well as reliable surveillance, is essential for case management and patient improvement.
Objectives: For this research, our aim was to evaluate the application of a novel SYBR Green based one-step real-time reverse-transcriptase polymerase chain reaction (rRT-PCR) assay for the in-house diagnosis of the CCHF virus.
Patients and methods: In this experimental study, the highly conserved S-region sequence of the CCHF viral genome was first adapted from GenBank, and the specific primers targeting this region were designed. Then, the viral RNA was extracted from 75 serum samples from different patients in eastern Iran. The sensitivity and specificity of the primers were also evaluated in positive serum samples previously confirmed to have the CCHF virus, by this one-step rRT-PCR assay, as well as a DNA sequencing analysis.
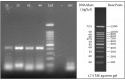
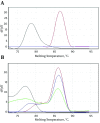
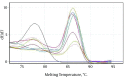
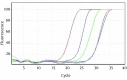
Results: From a total of 75 suspected serum samples, 42 were confirmed to be positive for CCHF virus, with no false-positives detected by the sequencing results. After 40 amplification cycles, the melting curve analysis revealed a mean melting temperature (Tm) of 86.5 ± 0.6°C (quite different from those of the primer-dimers), and the positive samples showed only a small variation in the parameters. In all of the positive samples, the predicted length of 420 bp was confirmed by electrophoresis. Moreover, the sensitivity test showed that this assay can detect less than 20 copies of viral RNA per reaction.
Conclusions: This study showed that this novel one-step rRT-PCR assay is a rapid, reliable, repeatable, specific, sensitive, and simple tool for the detection of the CCHF virus.
Keywords: Crimean-Congo; Diagnosis; Hemorrhagic Fever Virus; Real-Time Polymerase Chain Reaction.
Figures
Similar articles
-
Rapid and quantitative detection of Crimean-Congo hemorrhagic fever virus by one-step real-time reverse transcriptase-PCR.Jpn J Infect Dis. 2005 Dec;58(6):358-62. Jpn J Infect Dis. 2005. PMID: 16377867
-
Novel one-step real-time RT-PCR assay for rapid and specific diagnosis of Crimean-Congo hemorrhagic fever encountered in the Balkans.J Virol Methods. 2006 May;133(2):175-9. doi: 10.1016/j.jviromet.2005.11.006. Epub 2005 Dec 15. J Virol Methods. 2006. PMID: 16343650
-
Molecular and serological findings in suspected patients with Crimean-Congo hemorrhagic fever virus in Iran.J Med Virol. 2015 Apr;87(4):686-93. doi: 10.1002/jmv.24106. Epub 2015 Feb 3. J Med Virol. 2015. PMID: 25649667
-
Diagnosis of Crimean-Congo hemorrhagic fever.Expert Rev Anti Infect Ther. 2015 May;13(5):555-66. doi: 10.1586/14787210.2015.1021782. Epub 2015 Mar 6. Expert Rev Anti Infect Ther. 2015. PMID: 25746112 Review.
-
Crimean-Congo hemorrhagic fever.Antiviral Res. 2004 Dec;64(3):145-60. doi: 10.1016/j.antiviral.2004.08.001. Antiviral Res. 2004. PMID: 15550268 Review.
Cited by
-
A Novel RT-LAMP for the Detection of Different Genotypes of Crimean-Congo Haemorrhagic Fever Virus in Patients from Spain.Int J Mol Sci. 2023 Mar 29;24(7):6411. doi: 10.3390/ijms24076411. Int J Mol Sci. 2023. PMID: 37047384 Free PMC article.
-
Recent Advances in Crimean-Congo Hemorrhagic Fever Virus Detection, Treatment, and Vaccination: Overview of Current Status and Challenges.Biol Proced Online. 2024 Jun 26;26(1):20. doi: 10.1186/s12575-024-00244-3. Biol Proced Online. 2024. PMID: 38926669 Free PMC article. Review.
-
Diagnostic Testing for Crimean-Congo Hemorrhagic Fever.J Clin Microbiol. 2020 Mar 25;58(4):e01580-19. doi: 10.1128/JCM.01580-19. Print 2020 Mar 25. J Clin Microbiol. 2020. PMID: 32024724 Free PMC article. Review.
References
-
- Vazirianzadeh B, Rahdar M. Correct identification of animal host species is important in the diagnosis of Zoonotic diseases. Jundishapur J Microbiol. 2013;6(2):97–9. doi: 10.5812/jjm.9537. - DOI
-
- Drosten C, Gottig S, Schilling S, Asper M, Panning M, Schmitz H, et al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol. 2002;40(7):2323–30. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
