Primer Evaluation for PCR and its Application for Detection of Carbapenemases in Enterobacteriaceae
- PMID: 27099689
- PMCID: PMC4834133
- DOI: 10.5812/jjm.29314
Primer Evaluation for PCR and its Application for Detection of Carbapenemases in Enterobacteriaceae
Abstract
Background: During the last decade, the prevalence of carbapenem-resistant Enterobacteriaceae in human patients has increased. Carbapenemase-producing bacteria are usually multidrug resistant. Therefore, early recognition of carbapenemase producers is critical to prevent their spread.
Objectives: The objective of this study was to develop the primers for single and/or multiplex PCR amplification assays for simultaneous identification of class A, class B, and class D carbapenem hydrolyzing β-lactamases in Enterobacteriaceae and then to evaluate their efficiency.
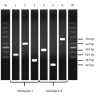
Materials and methods: The reference sequences of all genes encoding carbapenemases were downloaded from GenBank. Primers were designed to amplify the following 11 genes: bla KPC, bla OXA, bla VIM, bla NDM, bla IMP, bla SME, bla IMI, bla GES , bla GIM, bla DIM and bla CMY . PCR conditions were tested to amplify fragments of different sizes. Two multiplex PCR sets were created for the detection of clinically important carbapenemases. The third set of primers was included for detection of all known carbapenemases in Enterobacteriaceae. They were evaluated using six reference strains and nine clinical isolates.
Results: Using optimized conditions, all carbapenemase-positive controls yielded predicted amplicon sizes and confirmed the specificity of the primers in single and multiplex PCR.
Conclusions: We have reported here a reliable method, composed of single and multiplex PCR assays, for screening all clinically known carbapenemases. Primers tested in silico and in vitro may distinguish carbapenem-resistant Enterobacteriaceae and could assist in combating the spread of carbapenem resistance in Enterobacteriaceae.
Keywords: Carbapenems; Drug Resistance; Enterobacteriaceae; Multiplex Polymerase Chain Reaction.
Figures
References
-
- Mandell GL, Douglas RG, Bennett JE, Dolin R. Mandell, Douglas, and Bennett's principles and practice of infectious diseases. 6th ed. New York: Elsevier; 2005.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
