Migration and DNA methylation: a comparison of methylation patterns in type 2 diabetes susceptibility genes between indians and europeans
- PMID: 27099715
- PMCID: PMC4835020
- DOI: 10.7243/2050-0866-2-6
Migration and DNA methylation: a comparison of methylation patterns in type 2 diabetes susceptibility genes between indians and europeans
Abstract
Background: Type 2 diabetes is a global problem that is increasingly prevalent in low and middle income countries including India, and is partly attributed to increased urbanisation. Genotype clearly plays a role in type 2 diabetes susceptibility. However, the role of DNA methylation and its interaction with genotype and metabolic measures is poorly understood. This study aimed to establish whether methylation patterns of type 2 diabetes genes differ between distinct Indian and European populations and/or change following rural to urban migration in India.
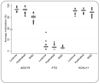
Methods: Quantitative DNA methylation analysis in Indians and Europeans using Sequenom® EpiTYPER® technology was undertaken in three genes: ADCY5, FTO and KCNJ11. Metabolic measures and genotype data were also analysed.
Results: Consistent differences in DNA methylation patterns were observed between Indian and European populations in ADCY5, FTO and KCNJ11. Associations were demonstrated between FTO rs9939609 and BMI and between ADCY5rs17295401 and HDL levels in Europeans. However, these observations were not linked to local variation in DNA methylation levels. No differences in methylation patterns were observed in urban-dwelling migrants compared to their non-migrant rural-dwelling siblings in India.
Conclusions: Analysis of DNA methylation at three type 2 diabetes susceptibility loci highlighted geographical and ethnic differences in methylation patterns. These differences may be attributed to genetic and/or region-specific environmental factors.
Keywords: DNA methylation; Type 2 diabetes; ethnicity.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
FTO gene variants are strongly associated with type 2 diabetes in South Asian Indians.Diabetologia. 2009 Feb;52(2):247-52. doi: 10.1007/s00125-008-1186-6. Epub 2008 Nov 13. Diabetologia. 2009. PMID: 19005641 Free PMC article.
-
Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study.Lancet Diabetes Endocrinol. 2015 Jul;3(7):526-534. doi: 10.1016/S2213-8587(15)00127-8. Epub 2015 Jun 18. Lancet Diabetes Endocrinol. 2015. PMID: 26095709 Free PMC article.
-
rs9939609 FTO genotype associations with FTO methylation level influences body mass and telomere length in an Australian rural population.Int J Obes (Lond). 2017 Sep;41(9):1427-1433. doi: 10.1038/ijo.2017.127. Epub 2017 May 31. Int J Obes (Lond). 2017. PMID: 28559540
-
Implications of critical PPARγ2, ADIPOQ and FTO gene polymorphisms in type 2 diabetes and obesity-mediated susceptibility to type 2 diabetes in an Indian population.Mol Genet Genomics. 2016 Feb;291(1):193-204. doi: 10.1007/s00438-015-1097-4. Epub 2015 Aug 5. Mol Genet Genomics. 2016. PMID: 26243686
-
Impact of Type 2 Diabetes Mellitus with a Focus on Asian Indians Living in India and Abroad: A Systematic Review.Endocr Metab Immune Disord Drug Targets. 2023;23(5):609-616. doi: 10.2174/1871530322666220827161236. Endocr Metab Immune Disord Drug Targets. 2023. PMID: 36043735
Cited by
-
Differences in smoking associated DNA methylation patterns in South Asians and Europeans.Clin Epigenetics. 2014 Feb 3;6(1):4. doi: 10.1186/1868-7083-6-4. Clin Epigenetics. 2014. PMID: 24485148 Free PMC article.
-
Type 2 diabetes is more closely associated with risk of colorectal cancer based on elevated DNA methylation levels of ADCY5.Oncol Lett. 2022 May 12;24(1):206. doi: 10.3892/ol.2022.13327. eCollection 2022 Jul. Oncol Lett. 2022. PMID: 35720494 Free PMC article.
-
New insights from monogenic diabetes for "common" type 2 diabetes.Front Genet. 2015 Aug 7;6:251. doi: 10.3389/fgene.2015.00251. eCollection 2015. Front Genet. 2015. PMID: 26300908 Free PMC article. Review.
-
Fetal programming of adipose tissue function: an evolutionary perspective.Mamm Genome. 2014 Oct;25(9-10):413-23. doi: 10.1007/s00335-014-9528-9. Epub 2014 Jun 27. Mamm Genome. 2014. PMID: 24969535 Review.
References
-
- Sandovici I, Smith NH, Nitert MD, Ackers-Johnson M, Uribe-Lewis S, Ito Y, Jones RH, Marquez VE, Cairns W, Tadayyon M, O’Neill LP, et al. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc Natl Acad Sci U S A. 2011;108:5449–54. - PMC - PubMed
-
- Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr. 2007;97:1064–73. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
