Germline Gene Editing in Chickens by Efficient CRISPR-Mediated Homologous Recombination in Primordial Germ Cells
- PMID: 27099923
- PMCID: PMC4839619
- DOI: 10.1371/journal.pone.0154303
Germline Gene Editing in Chickens by Efficient CRISPR-Mediated Homologous Recombination in Primordial Germ Cells
Abstract
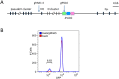
The CRISPR/Cas9 system has been applied in a large number of animal and plant species for genome editing. In chickens, CRISPR has been used to knockout genes in somatic tissues, but no CRISPR-mediated germline modification has yet been reported. Here we use CRISPR to target the chicken immunoglobulin heavy chain locus in primordial germ cells (PGCs) to produce transgenic progeny. Guide RNAs were co-transfected with a donor vector for homology-directed repair of the double-strand break, and clonal populations were selected. All of the resulting drug-resistant clones contained the correct targeting event. The targeted cells gave rise to healthy progeny containing the CRISPR-targeted locus. The results show that gene-edited chickens can be obtained by modifying PGCs in vitro with the CRISPR/Cas9 system, opening up many potential applications for efficient genetic modification in birds.
Conflict of interest statement
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

