PBRM1 Regulates the Expression of Genes Involved in Metabolism and Cell Adhesion in Renal Clear Cell Carcinoma
- PMID: 27100670
- PMCID: PMC4839679
- DOI: 10.1371/journal.pone.0153718
PBRM1 Regulates the Expression of Genes Involved in Metabolism and Cell Adhesion in Renal Clear Cell Carcinoma
Abstract
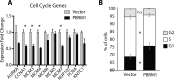
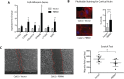
Polybromo-1 (PBRM1) is a component of the PBAF (Polybromo-associated-BRG1- or BRM-associated factors) chromatin remodeling complex and is the second most frequently mutated gene in clear-cell renal cell Carcinoma (ccRCC). Mutation of PBRM1 is believed to be an early event in carcinogenesis, however its function as a tumor suppressor is not understood. In this study, we have employed Next Generation Sequencing to profile the differentially expressed genes upon PBRM1 re-expression in a cellular model of ccRCC. PBRM1 re-expression led to upregulation of genes involved in cellular adhesion, carbohydrate metabolism, apoptotic process and response to hypoxia, and a downregulation of genes involved in different stages of cell division. The decrease in cellular proliferation upon PBRM1 re-expression was confirmed, validating the functional role of PBRM1 as a tumor suppressor in a cell-based model. In addition, we identified a role for PBRM1 in regulating metabolic pathways known to be important for driving ccRCC, including the regulation of hypoxia response genes, PI3K signaling, glucose uptake, and cholesterol homeostasis. Of particular novelty is the identification of cell adhesion as a major downstream process uniquely regulated by PBRM1 expression. Cytoskeletal reorganization was induced upon PBRM1 reexpression as evidenced from the increase in the number of cells displaying cortical actin, a hallmark of epithelial cells. Genes involved in cell adhesion featured prominently in our transcriptional dataset and overlapped with genes uniquely regulated by PBRM1 in clinical specimens of ccRCC. Genes involved in cell adhesion serve as tumor suppressor and maybe involved in inhibiting cell migration. Here we report for the first time genes linked to cell adhesion serve as downstream targets of PBRM1, and hope to lay the foundation of future studies focusing on the role of chromatin remodelers in bringing about these alterations during malignancies.
Conflict of interest statement
Figures





References
-
- Motzer RJ, Jonasch E, Agarwal N, Beard C, Bhayani S, Bolger GB, et al. Kidney cancer, version 3.2015. Journal Of The National Comprehensive Cancer Network: JNCCN. 2015;13(2):151 - PubMed
-
- Duns G, Hofstra RM, Sietzema JG, Hollema H, van Duivenbode I, Kuik A, et al. Targeted exome sequencing in clear cell renal cell carcinoma tumors suggests aberrant chromatin regulation as a crucial step in ccRCC development. Hum Mutat. 2012;33(7):1059–62. Epub 2012/03/31. 10.1002/humu.22090 . - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

