CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing
- PMID: 27100738
- PMCID: PMC4839673
- DOI: 10.1371/journal.pcbi.1004873
CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing
Abstract
Germline copy number variants (CNVs) and somatic copy number alterations (SCNAs) are of significant importance in syndromic conditions and cancer. Massively parallel sequencing is increasingly used to infer copy number information from variations in the read depth in sequencing data. However, this approach has limitations in the case of targeted re-sequencing, which leaves gaps in coverage between the regions chosen for enrichment and introduces biases related to the efficiency of target capture and library preparation. We present a method for copy number detection, implemented in the software package CNVkit, that uses both the targeted reads and the nonspecifically captured off-target reads to infer copy number evenly across the genome. This combination achieves both exon-level resolution in targeted regions and sufficient resolution in the larger intronic and intergenic regions to identify copy number changes. In particular, we successfully inferred copy number at equivalent to 100-kilobase resolution genome-wide from a platform targeting as few as 293 genes. After normalizing read counts to a pooled reference, we evaluated and corrected for three sources of bias that explain most of the extraneous variability in the sequencing read depth: GC content, target footprint size and spacing, and repetitive sequences. We compared the performance of CNVkit to copy number changes identified by array comparative genomic hybridization. We packaged the components of CNVkit so that it is straightforward to use and provides visualizations, detailed reporting of significant features, and export options for integration into existing analysis pipelines. CNVkit is freely available from https://github.com/etal/cnvkit.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
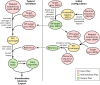
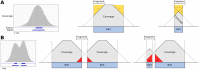
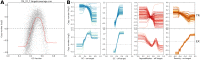
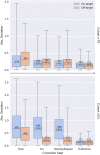


References
-
- Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, et al. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proceedings of the National Academy of Sciences of the United States of America. 2007. May;104(22):9387–92. 10.1073/pnas.0702165104 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

