A Balanced Tissue Composition Reveals New Metabolic and Gene Expression Markers in Prostate Cancer
- PMID: 27100877
- PMCID: PMC4839647
- DOI: 10.1371/journal.pone.0153727
A Balanced Tissue Composition Reveals New Metabolic and Gene Expression Markers in Prostate Cancer
Abstract
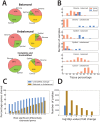
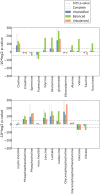
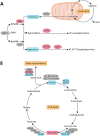
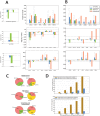
Molecular analysis of patient tissue samples is essential to characterize the in vivo variability in human cancers which are not accessible in cell-lines or animal models. This applies particularly to studies of tumor metabolism. The challenge is, however, the complex mixture of various tissue types within each sample, such as benign epithelium, stroma and cancer tissue, which can introduce systematic biases when cancers are compared to normal samples. In this study we apply a simple strategy to remove such biases using sample selections where the average content of stroma tissue is balanced between the sample groups. The strategy is applied to a prostate cancer patient cohort where data from MR spectroscopy and gene expression have been collected from and integrated on the exact same tissue samples. We reveal in vivo changes in cancer-relevant metabolic pathways which are otherwise hidden in the data due to tissue confounding. In particular, lowered levels of putrescine are connected to increased expression of SRM, reduced levels of citrate are attributed to upregulation of genes promoting fatty acid synthesis, and increased succinate levels coincide with reduced expression of SUCLA2 and SDHD. In addition, the strategy also highlights important metabolic differences between the stroma, epithelium and prostate cancer. These results show that important in vivo metabolic features of cancer can be revealed from patient data only if the heterogeneous tissue composition is properly accounted for in the analysis.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

