Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome
- PMID: 27102203
- PMCID: PMC4841045
- DOI: 10.1186/s13073-016-0293-0
Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome
Abstract
Background: The oral cavity is home to one of the most diverse microbial communities of the human body and a major entry portal for pathogens. Its homeostasis is maintained by saliva, which fulfills key functions including lubrication of food, pre-digestion, and bacterial defense. Consequently, disruptions in saliva secretion and changes in the oral microbiome contribute to conditions such as tooth decay and respiratory tract infections. Here we set out to quantitatively map the saliva proteome in great depth with a rapid and in-depth mass spectrometry-based proteomics workflow.
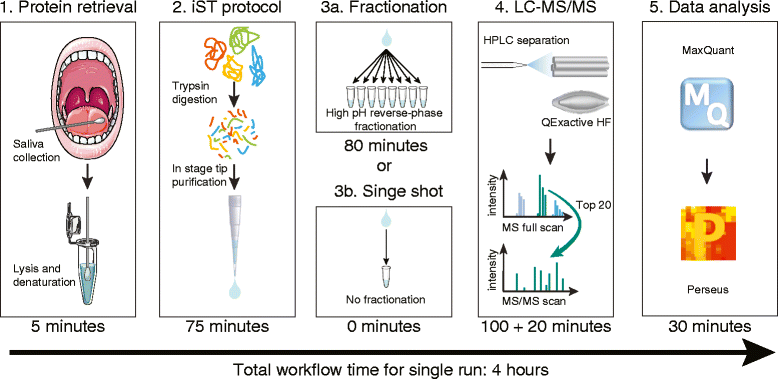
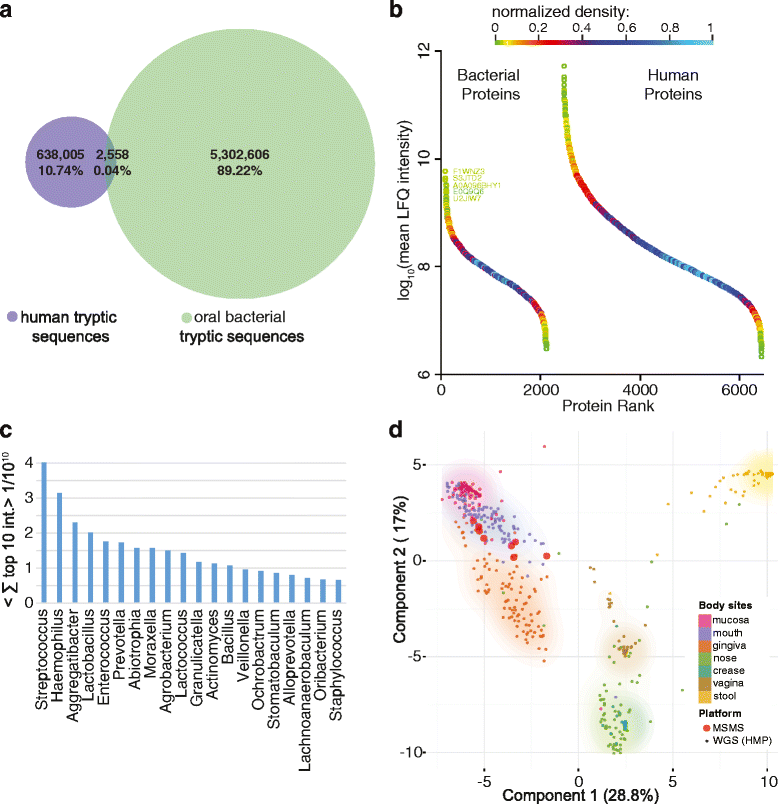
Methods: We used recent improvements in mass spectrometry (MS)-based proteomics to develop a rapid workflow for mapping the saliva proteome quantitatively and at great depth. Standard clinical cotton swabs were used to collect saliva form eight healthy individuals at two different time points, allowing us to study inter-individual differences and interday changes of the saliva proteome. To accurately identify microbial proteins, we developed a method called "split by taxonomy id" that prevents peptides shared by humans and bacteria or between different bacterial phyla to contribute to protein identification.
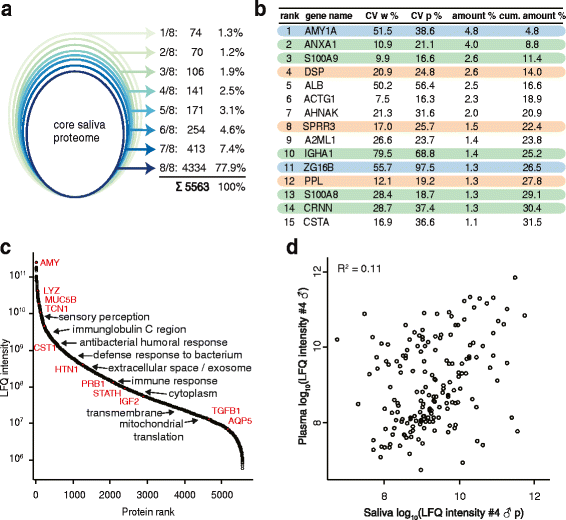
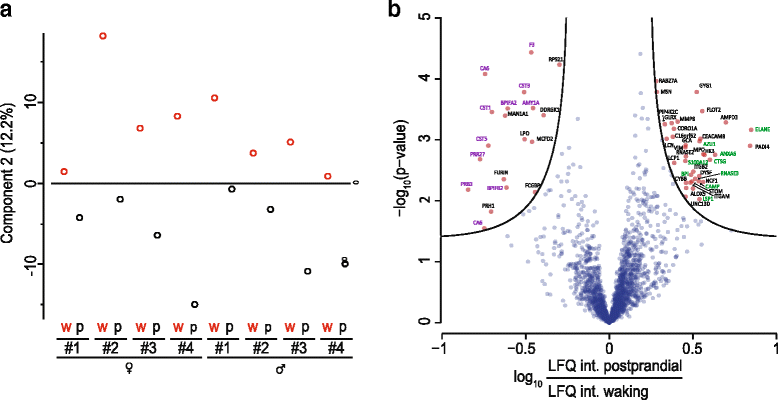
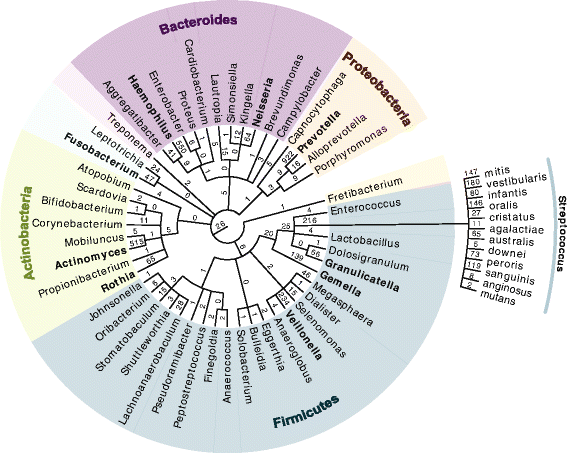
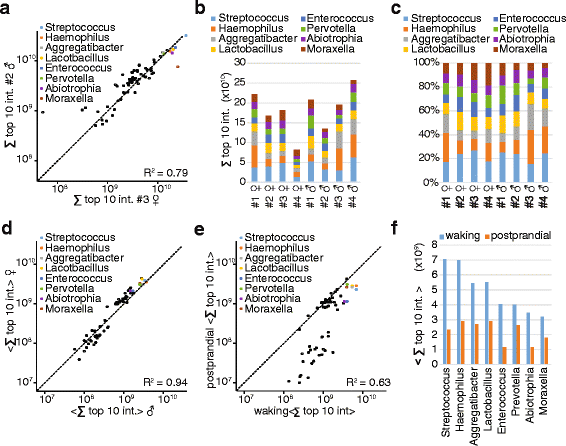
Results: Microgram protein amounts retrieved from cotton swabs resulted in more than 3700 quantified human proteins in 100-min gradients or 5500 proteins after simple fractionation. Remarkably, our measurements also quantified more than 2000 microbial proteins from 50 bacterial genera. Co-analysis of the proteomics results with next-generation sequencing data from the Human Microbiome Project as well as a comparison to MALDI-TOF mass spectrometry on microbial cultures revealed strong agreement. The oral microbiome differs between individuals and changes drastically upon eating and tooth brushing.
Conclusion: Rapid shotgun and robust technology can now simultaneously characterize the human and microbiome contributions to the proteome of a body fluid and is therefore a valuable complement to genomic studies. This opens new frontiers for the study of host-pathogen interactions and clinical saliva diagnostics.
Figures
References
-
- Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. J Proteome Res. 2009;8:5590–600. doi: 10.1021/pr900675w. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

