An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis
- PMID: 27102666
- PMCID: PMC4840970
- DOI: 10.1186/s13073-016-0299-7
An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis
Abstract
Background: The adaptive immune response in rheumatoid arthritis (RA) is influenced by an interaction between host genetics and environment, particularly the host microbiome. Association of the gut microbiota with various diseases has been reported, though the specific components of the microbiota that affect the host response leading to disease remain unknown. However, there is limited information on the role of gut microbiota in RA. In this study we aimed to define a microbial and metabolite profile that could predict disease status. In addition, we aimed to generate a humanized model of arthritis to confirm the RA-associated microbe.
Methods: To identify an RA biomarker profile, the 16S ribosomal DNA of fecal samples from RA patients, first-degree relatives (to rule out environment/background as confounding factors), and random healthy non-RA controls were sequenced. Analysis of metabolites and their association with specific taxa was performed to investigate a potential mechanistic link. The role of an RA-associated microbe was confirmed using a human epithelial cell line and a humanized mouse model of arthritis.
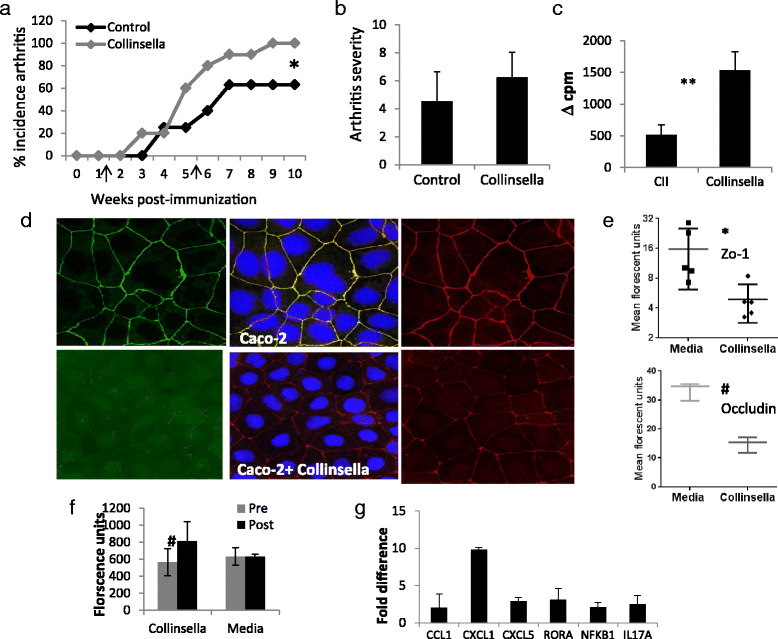
Results: Patients with RA exhibited decreased gut microbial diversity compared with controls, which correlated with disease duration and autoantibody levels. A taxon-level analysis suggested an expansion of rare taxa, Actinobacteria, with a decrease in abundant taxa in patients with RA compared with controls. Prediction models based on the random forests algorithm suggested that three genera, Collinsella, Eggerthella, and Faecalibacterium, segregated with RA. The abundance of Collinsella correlated strongly with high levels of alpha-aminoadipic acid and asparagine as well as production of the proinflammatory cytokine IL-17A. A role for Collinsella in altering gut permeability and disease severity was confirmed in experimental arthritis.
Conclusions: These observations suggest dysbiosis in RA patients resulting from the abundance of certain rare bacterial lineages. A correlation between the intestinal microbiota and metabolic signatures could determine a predictive profile for disease causation and progression.
Figures





References
-
- Silman AJ, MacGregor AJ, Thomson W, Holligan S, Carthy D, Farhan A, Ollier WE. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br J Rheumatol. 1993;32:903–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

