Comprehensive Molecular Characterization of Salivary Duct Carcinoma Reveals Actionable Targets and Similarity to Apocrine Breast Cancer
- PMID: 27103403
- PMCID: PMC5026550
- DOI: 10.1158/1078-0432.CCR-16-0637
Comprehensive Molecular Characterization of Salivary Duct Carcinoma Reveals Actionable Targets and Similarity to Apocrine Breast Cancer
Abstract
Purpose: Salivary duct carcinoma (SDC) is an aggressive salivary malignancy, which is resistant to chemotherapy and has high mortality rates. We investigated the molecular landscape of SDC, focusing on genetic alterations and gene expression profiles.
Experimental design: We performed whole-exome sequencing, RNA sequencing, and immunohistochemical analyses in 16 SDC tumors and examined selected alterations via targeted sequencing of 410 genes in a second cohort of 15 SDCs.
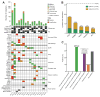
Results: SDCs harbored a higher mutational burden than many other salivary carcinomas (1.7 mutations/Mb). The most frequent genetic alterations were mutations in TP53 (55%), HRAS (23%), PIK3CA (23%), and amplification of ERBB2 (35%). Most (74%) tumors had alterations in either MAPK (BRAF/HRAS/NF1) genes or ERBB2 Potentially targetable alterations based on supportive clinical evidence were present in 61% of tumors. Androgen receptor (AR) was overexpressed in 75%; several potential resistance mechanisms to androgen deprivation therapy (ADT) were identified, including the AR-V7 splice variant (present in 50%, often at low ratios compared with full-length AR) and FOXA1 mutations (10%). Consensus clustering and pathway analyses in transcriptome data revealed striking similarities between SDC and molecular apocrine breast cancer.
Conclusions: This study illuminates the landscape of genetic alterations and gene expression programs in SDC, identifying numerous molecular targets and potential determinants of response to AR antagonism. This has relevance for emerging clinical studies of ADT and other targeted therapies in SDC. The similarities between SDC and apocrine breast cancer indicate that clinical data in breast cancer may generate useful hypotheses for SDC. Clin Cancer Res; 22(18); 4623-33. ©2016 AACR.
©2016 American Association for Cancer Research.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Luk PP, Weston JD, Yu B, Selinger CI, Ekmejian R, Eviston TJ, et al. Salivary duct carcinoma: Clinicopathologic features, morphologic spectrum, and somatic mutations. Head Neck. 2015 - PubMed
-
- Chiosea SI, Williams L, Griffith CC, Thompson LD, Weinreb I, Bauman JE, et al. Molecular characterization of apocrine salivary duct carcinoma. Am J Surg Pathol. 2015;39(6):744–52. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

