Integrated Genomics Reveals Convergent Transcriptomic Networks Underlying Chronic Obstructive Pulmonary Disease and Idiopathic Pulmonary Fibrosis
- PMID: 27104832
- PMCID: PMC5067817
- DOI: 10.1164/rccm.201510-2026OC
Integrated Genomics Reveals Convergent Transcriptomic Networks Underlying Chronic Obstructive Pulmonary Disease and Idiopathic Pulmonary Fibrosis
Abstract
Rationale: Despite shared environmental exposures, idiopathic pulmonary fibrosis (IPF) and chronic obstructive pulmonary disease are usually studied in isolation, and the presence of shared molecular mechanisms is unknown.
Objectives: We applied an integrative genomic approach to identify convergent transcriptomic pathways in emphysema and IPF.
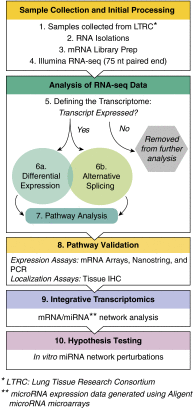
Methods: We defined the transcriptional repertoire of chronic obstructive pulmonary disease, IPF, or normal histology lungs using RNA-seq (n = 87).
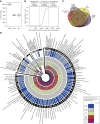
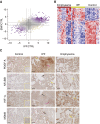
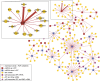
Measurements and main results: Genes increased in both emphysema and IPF relative to control were enriched for the p53/hypoxia pathway, a finding confirmed in an independent cohort using both gene expression arrays and the nCounter Analysis System (n = 193). Immunohistochemistry confirmed overexpression of HIF1A, MDM2, and NFKBIB members of this pathway in tissues from patients with emphysema or IPF. Using reads aligned across splice junctions, we determined that alternative splicing of p53/hypoxia pathway-associated molecules NUMB and PDGFA occurred more frequently in IPF or emphysema compared with control and validated these findings by quantitative polymerase chain reaction and the nCounter Analysis System on an independent sample set (n = 193). Finally, by integrating parallel microRNA and mRNA-Seq data on the same samples, we identified MIR96 as a key novel regulatory hub in the p53/hypoxia gene-expression network and confirmed that modulation of MIR96 in vitro recapitulates the disease-associated gene-expression network.
Conclusions: Our results suggest convergent transcriptional regulatory hubs in diseases as varied phenotypically as chronic obstructive pulmonary disease and IPF and suggest that these hubs may represent shared key responses of the lung to environmental stresses.
Keywords: COPD; ILD; IPF; network; transcriptome.
Figures

Comment in
-
Biological Network Modeling and Systems Biology to Advance Our Understanding of Lung Disease.Am J Respir Crit Care Med. 2016 Oct 15;194(8):920-921. doi: 10.1164/rccm.201604-0793ED. Am J Respir Crit Care Med. 2016. PMID: 27739892 No abstract available.
References
-
- Mannino DM, Homa DM, Akinbami LJ, Ford ES, Redd SC. Chronic obstructive pulmonary disease surveillance—United States, 1971-2000. Respir Care. 2002;47:1184–1199. - PubMed
-
- Selman M, King TE, Pardo A American Thoracic Society; European Respiratory Society; American College of Chest Physicians. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med. 2001;134:136–151. - PubMed
-
- Pauwels RA, Buist AS, Calverley PMA, Jenkins CR, Hurd SS GOLD Scientific Committee. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001;163:1256–1276. - PubMed
-
- Wang I-M, Stepaniants S, Boie Y, Mortimer JR, Kennedy B, Elliott M, Hayashi S, Loy L, Coulter S, Cervino S, et al. Gene expression profiling in patients with chronic obstructive pulmonary disease and lung cancer. Am J Respir Crit Care Med. 2008;177:402–411. - PubMed
-
- Spira A, Beane J, Pinto-Plata V, Kadar A, Liu G, Shah V, Celli B, Brody JS. Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol. 2004;31:601–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

