Methylome-wide Analysis of Chronic HIV Infection Reveals Five-Year Increase in Biological Age and Epigenetic Targeting of HLA
- PMID: 27105112
- PMCID: PMC4995115
- DOI: 10.1016/j.molcel.2016.03.019
Methylome-wide Analysis of Chronic HIV Infection Reveals Five-Year Increase in Biological Age and Epigenetic Targeting of HLA
Abstract
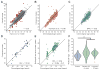
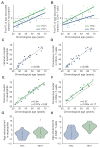
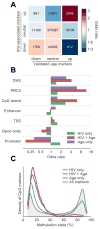
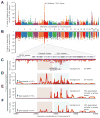
HIV-infected individuals are living longer on antiretroviral therapy, but many patients display signs that in some ways resemble premature aging. To investigate and quantify the impact of chronic HIV infection on aging, we report a global analysis of the whole-blood DNA methylomes of 137 HIV+ individuals under sustained therapy along with 44 matched HIV- individuals. First, we develop and validate epigenetic models of aging that are independent of blood cell composition. Using these models, we find that both chronic and recent HIV infection lead to an average aging advancement of 4.9 years, increasing expected mortality risk by 19%. In addition, sustained infection results in global deregulation of the methylome across >80,000 CpGs and specific hypomethylation of the region encoding the human leukocyte antigen locus (HLA). We find that decreased HLA methylation is predictive of lower CD4 / CD8 T cell ratio, linking molecular aging, epigenetic regulation, and disease progression.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

Comment in
-
An Epigenetic Clock Measures Accelerated Aging in Treated HIV Infection.Mol Cell. 2016 Apr 21;62(2):153-155. doi: 10.1016/j.molcel.2016.04.008. Mol Cell. 2016. PMID: 27105110 Free PMC article.
References
-
- Althoff KN, McGinnis KA, Wyatt CM, Freiberg MS, Gilbert C, Oursler KK, Rimland D, Rodriguez-Barradas MC, Dubrow R, Park LS, et al. Comparison of Risk and Age at Diagnosis of Myocardial Infarction, End-Stage Renal Disease, and Non-AIDS-Defining Cancer in HIV-Infected Versus Uninfected Adults. Clin Infect Dis 2014 - PMC - PubMed
-
- Appay V, Rowland-Jones SL. Premature ageing of the immune system: the cause of AIDS? Trends Immunol. 2002;23:580–585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

