Monitoring the spread of meticillin-resistant Staphylococcus aureus in The Netherlands from a reference laboratory perspective
- PMID: 27105754
- PMCID: PMC4964845
- DOI: 10.1016/j.jhin.2016.02.022
Monitoring the spread of meticillin-resistant Staphylococcus aureus in The Netherlands from a reference laboratory perspective
Abstract
Background: In The Netherlands, efforts to control meticillin-resistant Staphylococcus aureus (MRSA) in hospitals have been largely successful due to stringent screening of patients on admission and isolation of those that fall into defined risk categories. However, Dutch hospitals are not free of MRSA, and a considerable number of cases are found that do not belong to any of the risk categories. Some of these may be due to undetected nosocomial transmission, whereas others may be introduced from unknown reservoirs.
Aim: Identifying multi-institutional clusters of MRSA isolates to estimate the contribution of potential unobserved reservoirs in The Netherlands.
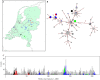
Methods: We applied a clustering algorithm that combines time, place, and genetics to routine data available for all MRSA isolates submitted to the Dutch Staphylococcal Reference Laboratory between 2008 and 2011 in order to map the geo-temporal distribution of MRSA clonal lineages in The Netherlands.
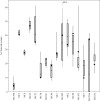
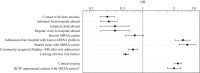
Findings: Of the 2966 isolates lacking obvious risk factors, 579 were part of geo-temporal clusters, whereas 2387 were classified as MRSA of unknown origin (MUOs). We also observed marked differences in the proportion of isolates that belonged to geo-temporal clusters between specific multi-locus variable number of tandem repeat analysis (MLVA) clonal complexes, indicating lineage-specific transmissibility. The majority of clustered isolates (74%) were present in multi-institutional clusters.
Conclusion: The frequency of MRSA of unknown origin among patients lacking obvious risk factors is an indication of a largely undefined extra-institutional but genetically highly diverse reservoir. Efforts to understand the emergence and spread of high-risk clones require the pooling of routine epidemiological information and typing data into central databases.
Keywords: Algorithm; Epidemiology; Meticillin-resistant Staphylococcus aureus; Surveillance.
Copyright © 2016. Published by Elsevier Ltd.
Figures

References
-
- EARS-Net, European Antimicrobial Resistance Surveillance Network. Annual report, 2011. p. 55–58. Available at: http://www.ecdc.europa.eu/en/publications/Publications/antimicrobial-res... [last accessed March 2016].
-
- Vandenbroucke-Grauls C.M. Methicillin-resistant Staphylococcus aureus control in hospitals: the Dutch experience. Infect Control Hosp Epidemiol. 1996;17:512–513. - PubMed
-
- Infection Prevention Working Party, Infection Prevention Working Party (WIP). Hospital MRSA guideline, 2012. Available at: http://www.wip.nl/UK/free_content/Richtlijnen/MRSA%20hospital.pdf [last accessed March 2016].
-
- Lekkerkerk W.S., van de Sande-Bruinsma N., van der Sande M.A. Emergence of MRSA of unknown origin in the Netherlands. Clin Microbiol Infect. 2012;18:656–661. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

