Hypermethylated in cancer 1(HIC1) suppresses non-small cell lung cancer progression by targeting interleukin-6/Stat3 pathway
- PMID: 27107418
- PMCID: PMC5058685
- DOI: 10.18632/oncotarget.8734
Hypermethylated in cancer 1(HIC1) suppresses non-small cell lung cancer progression by targeting interleukin-6/Stat3 pathway
Abstract
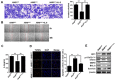
Non-small cell lung cancer (NSCLC), which accounts for more than 80% of lung cancers, is a leading cause of cancer mortality worldwide. However, the mechanism underlying its progression remains unclear. Here we found that HIC1 promoter was heavily methylated in NSCLC cell lines and tissues contributing to its low expression compared to normal controls. Restoring HIC1 expression inhibited migration, invasion and promoted inducible apoptosis of NSCLC cells. Notably, HIC1 is a tumor suppressor through inhibiting the transcription of IL-6 by sequence-specific binding on its promoter. Restoring IL-6 expression could partially rescue these phenotypes induced by HIC1 in vitro and in vivo. Mechanistic analyses show that autocrine secretion of IL-6 induced by loss of HIC1 activated STAT3 through IL-6/JAK pathway and was associated with NSCLC progression. The HIC1/IL-6 axis may serve as a prognostic biomarker and provide an attractive therapeutic target for NSCLC.
Keywords: HIC1/IL-6 axis; NSCLC; hypermethylation.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures

References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359–386. - PubMed
-
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA. 2014;64:9–29. - PubMed
-
- Temel JS, Greer JA, Muzikansky A, Gallagher ER, Admane S, Jackson VA, Dahlin CM, Blinderman CD, Jacobsen J, Pirl WF, Billings JA, Lynch TJ. Early palliative care for patients with metastatic non-small-cell lung cancer. The New England journal of medicine. 2010;363:733–742. - PubMed
-
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. The New England journal of medicine. 2003;349:2042–2054. - PubMed
-
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nature reviews Cancer. 2006;6:107–116. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

