Electronic medical record phenotyping using the anchor and learn framework
- PMID: 27107443
- PMCID: PMC4926745
- DOI: 10.1093/jamia/ocw011
Electronic medical record phenotyping using the anchor and learn framework
Abstract
Background: Electronic medical records (EMRs) hold a tremendous amount of information about patients that is relevant to determining the optimal approach to patient care. As medicine becomes increasingly precise, a patient's electronic medical record phenotype will play an important role in triggering clinical decision support systems that can deliver personalized recommendations in real time. Learning with anchors presents a method of efficiently learning statistically driven phenotypes with minimal manual intervention.
Materials and methods: We developed a phenotype library that uses both structured and unstructured data from the EMR to represent patients for real-time clinical decision support. Eight of the phenotypes were evaluated using retrospective EMR data on emergency department patients using a set of prospectively gathered gold standard labels.
Results: We built a phenotype library with 42 publicly available phenotype definitions. Using information from triage time, the phenotype classifiers have an area under the ROC curve (AUC) of infection 0.89, cancer 0.88, immunosuppressed 0.85, septic shock 0.93, nursing home 0.87, anticoagulated 0.83, cardiac etiology 0.89, and pneumonia 0.90. Using information available at the time of disposition from the emergency department, the AUC values are infection 0.91, cancer 0.95, immunosuppressed 0.90, septic shock 0.97, nursing home 0.91, anticoagulated 0.94, cardiac etiology 0.92, and pneumonia 0.97.
Discussion: The resulting phenotypes are interpretable and fast to build, and perform comparably to statistically learned phenotypes developed with 5000 manually labeled patients.
Conclusion: Learning with anchors is an attractive option for building a large public repository of phenotype definitions that can be used for a range of health IT applications, including real-time decision support.
Keywords: clinical decision support systems; electronic health records; knowledge representation; machine learning; natural language processing.
© The Author 2016. Published by Oxford University Press on behalf of the American Medical Informatics Association.
Figures

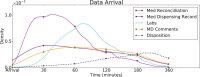

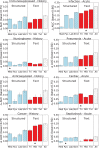
References
-
- Gandhi TK, Zuccotti G, Lee TH. Incomplete Care — On the Trail of Flaws in the System. New Engl J Med. 2011;365(6):486–488. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

