Molecular Mechanisms of Transcription Initiation-Structure, Function, and Evolution of TFE/TFIIE-Like Factors and Open Complex Formation
- PMID: 27107643
- PMCID: PMC7616663
- DOI: 10.1016/j.jmb.2016.04.016
Molecular Mechanisms of Transcription Initiation-Structure, Function, and Evolution of TFE/TFIIE-Like Factors and Open Complex Formation
Abstract
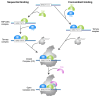
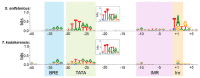
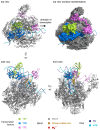
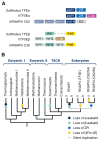
Transcription initiation requires that the promoter DNA is melted and the template strand is loaded into the active site of the RNA polymerase (RNAP), forming the open complex (OC). The archaeal initiation factor TFE and its eukaryotic counterpart TFIIE facilitate this process. Recent structural and biophysical studies have revealed the position of TFE/TFIIE within the pre-initiation complex (PIC) and illuminated its role in OC formation. TFE operates via allosteric and direct mechanisms. Firstly, it interacts with the RNAP and induces the opening of the flexible RNAP clamp domain, concomitant with DNA melting and template loading. Secondly, TFE binds physically to single-stranded DNA in the transcription bubble of the OC and increases its stability. The identification of the β-subunit of archaeal TFE enabled us to reconstruct the evolutionary history of TFE/TFIIE-like factors, which is characterised by winged helix (WH) domain expansion in eukaryotes and loss of metal centres including iron-sulfur clusters and Zinc ribbons. OC formation is an important target for the regulation of transcription in all domains of life. We propose that TFE and the bacterial general transcription factor CarD, although structurally and evolutionary unrelated, show interesting parallels in their mechanism to enhance OC formation. We argue that OC formation is used as a way to regulate transcription in all domains of life, and these regulatory mechanisms coevolved with the basal transcription machinery.
Keywords: RNA polymerase; TFIIE/TFE; archaea; evolution; transcription.
Copyright © 2016. Published by Elsevier Ltd.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

