Coupling of RNA Polymerase II Transcription Elongation with Pre-mRNA Splicing
- PMID: 27107644
- PMCID: PMC4893998
- DOI: 10.1016/j.jmb.2016.04.017
Coupling of RNA Polymerase II Transcription Elongation with Pre-mRNA Splicing
Abstract
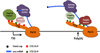
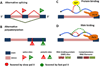
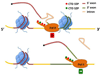
Pre-mRNA maturation frequently occurs at the same time and place as transcription by RNA polymerase II. The co-transcriptionality of mRNA processing has permitted the evolution of mechanisms that functionally couple transcription elongation with diverse events that occur on the nascent RNA. This review summarizes the current understanding of the relationship between transcriptional elongation through a chromatin template and co-transcriptional splicing including alternative splicing decisions that affect the expression of most human genes.
Keywords: CTD; RNA polymerase II; alternative splicing; kinetic coupling; transcription elongation.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
References
-
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

