Recessive and Dominant De Novo ITPR1 Mutations Cause Gillespie Syndrome
- PMID: 27108797
- PMCID: PMC4863566
- DOI: 10.1016/j.ajhg.2016.03.004
Recessive and Dominant De Novo ITPR1 Mutations Cause Gillespie Syndrome
Abstract
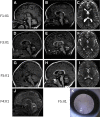
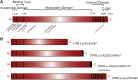
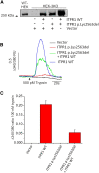
Gillespie syndrome (GS) is a rare variant form of aniridia characterized by non-progressive cerebellar ataxia, intellectual disability, and iris hypoplasia. Unlike the more common dominant and sporadic forms of aniridia, there has been no significant association with PAX6 mutations in individuals with GS and the mode of inheritance of the disease had long been regarded as uncertain. Using a combination of trio-based whole-exome sequencing and Sanger sequencing in five simplex GS-affected families, we found homozygous or compound heterozygous truncating mutations (c.4672C>T [p.Gln1558(∗)], c.2182C>T [p.Arg728(∗)], c.6366+3A>T [p.Gly2102Valfs5(∗)], and c.6664+5G>T [p.Ala2221Valfs23(∗)]) and de novo heterozygous mutations (c.7687_7689del [p.Lys2563del] and c.7659T>G [p.Phe2553Leu]) in the inositol 1,4,5-trisphosphate receptor type 1 gene (ITPR1). ITPR1 encodes one of the three members of the IP3-receptors family that form Ca(2+) release channels localized predominantly in membranes of endoplasmic reticulum Ca(2+) stores. The truncation mutants, which encompass the IP3-binding domain and varying lengths of the modulatory domain, did not form functional channels when produced in a heterologous cell system. Furthermore, ITPR1 p.Lys2563del mutant did not form IP3-induced Ca(2+) channels but exerted a negative effect when co-produced with wild-type ITPR1 channel activity. In total, these results demonstrate biallelic and monoallelic ITPR1 mutations as the underlying genetic defects for Gillespie syndrome, further extending the spectrum of ITPR1-related diseases.
Copyright © 2016 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Gillespie F.D. Aniridia, cerebellar ataxia, and oligophrenia in siblings. Arch. Ophthalmol. 1965;73:338–341. - PubMed
-
- Nelson J., Flaherty M., Grattan-Smith P. Gillespie syndrome: a report of two further cases. Am. J. Med. Genet. 1997;71:134–138. - PubMed
-
- François J., Lentini F. [Gillespie syndrome (incomplete aniridia, cerebellar ataxia and oligophrenia)] Klin. Monatsbl. Augenheilkd. 1984;184:313–315. - PubMed
-
- Wittig E.O., Moreira C.A., Freire-Maia N., Vianna-Morgante A.M. Partial aniridia, cerebellar ataxia, and mental deficiency (Gillespie syndrome) in two brothers. Am. J. Med. Genet. 1988;30:703–708. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

