A genetic basis for the variation in the vulnerability of cancer to DNA damage
- PMID: 27109210
- PMCID: PMC4848553
- DOI: 10.1038/ncomms11428
A genetic basis for the variation in the vulnerability of cancer to DNA damage
Abstract
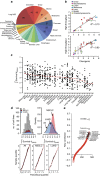
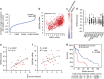
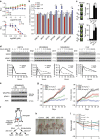
Radiotherapy is not currently informed by the genetic composition of an individual patient's tumour. To identify genetic features regulating survival after DNA damage, here we conduct large-scale profiling of cellular survival after exposure to radiation in a diverse collection of 533 genetically annotated human tumour cell lines. We show that sensitivity to radiation is characterized by significant variation across and within lineages. We combine results from our platform with genomic features to identify parameters that predict radiation sensitivity. We identify somatic copy number alterations, gene mutations and the basal expression of individual genes and gene sets that correlate with the radiation survival, revealing new insights into the genetic basis of tumour cellular response to DNA damage. These results demonstrate the diversity of tumour cellular response to ionizing radiation and establish multiple lines of evidence that new genetic features regulating cellular response after DNA damage can be identified.
Figures



References
-
- Alhassani A., Chandra A. & Chernew M. E. The sources of the SGR ‘hole'. N. Engl. J. Med. 366, 289–291 (2012). - PubMed
-
- Yard B., Chie E. K., Adams D. J., Peacock C. & Abazeed M. E. Radiotherapy in the era of precision medicine. Semin. Radiat. Oncol. 25, 227–236 (2015). - PubMed
-
- Lynch T. J. et al.. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 350, 2129–2139 (2004). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

