Mechanical cell competition kills cells via induction of lethal p53 levels
- PMID: 27109213
- PMCID: PMC4848481
- DOI: 10.1038/ncomms11373
Mechanical cell competition kills cells via induction of lethal p53 levels
Abstract
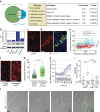
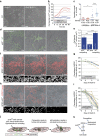
Cell competition is a quality control mechanism that eliminates unfit cells. How cells compete is poorly understood, but it is generally accepted that molecular exchange between cells signals elimination of unfit cells. Here we report an orthogonal mechanism of cell competition, whereby cells compete through mechanical insults. We show that MDCK cells silenced for the polarity gene scribble (scrib(KD)) are hypersensitive to compaction, that interaction with wild-type cells causes their compaction and that crowding is sufficient for scrib(KD) cell elimination. Importantly, we show that elevation of the tumour suppressor p53 is necessary and sufficient for crowding hypersensitivity. Compaction, via activation of Rho-associated kinase (ROCK) and the stress kinase p38, leads to further p53 elevation, causing cell death. Thus, in addition to molecules, cells use mechanical means to compete. Given the involvement of p53, compaction hypersensitivity may be widespread among damaged cells and offers an additional route to eliminate unfit cells.
Figures



References
-
- Morata G. & Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev. Biol. 42, 211–221 (1975). - PubMed
-
- Moreno E. & Basler K. dMyc transforms cells into super-competitors. Cell 117, 117–129 (2004). - PubMed
-
- la Cova, de C., Abril M., Bellosta P., Gallant P. & Johnston L. A. Drosophila myc regulates organ size by inducing cell competition. Cell 117, 107–116 (2004). - PubMed
-
- Oertel M., Menthena A., Dabeva M. D. & Shafritz D. A. Cell competition leads to a high level of normal liver reconstitution by transplanted fetal liver stem/progenitor cells. Gastroenterology 130, 507–520 (2006). - PubMed
-
- Rhiner C. et al.. Persistent competition among stem cells and their daughters in the Drosophila ovary germline niche. Development 136, 995–1006 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

