Sparse RNA folding revisited: space-efficient minimum free energy structure prediction
- PMID: 27110275
- PMCID: PMC4842305
- DOI: 10.1186/s13015-016-0071-y
Sparse RNA folding revisited: space-efficient minimum free energy structure prediction
Abstract
Background: RNA secondary structure prediction by energy minimization is the central computational tool for the analysis of structural non-coding RNAs and their interactions. Sparsification has been successfully applied to improve the time efficiency of various structure prediction algorithms while guaranteeing the same result; however, for many such folding problems, space efficiency is of even greater concern, particularly for long RNA sequences. So far, space-efficient sparsified RNA folding with fold reconstruction was solved only for simple base-pair-based pseudo-energy models.
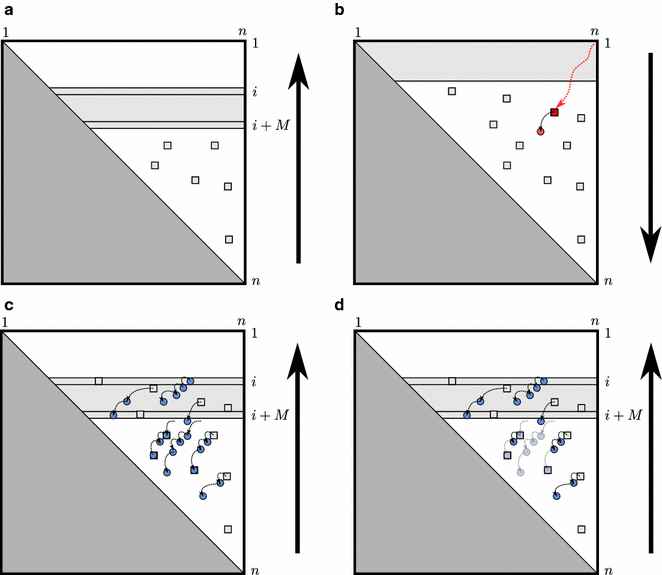
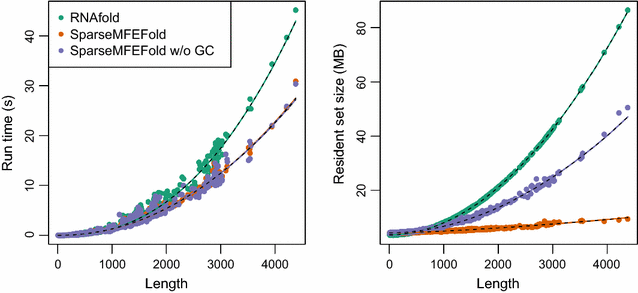
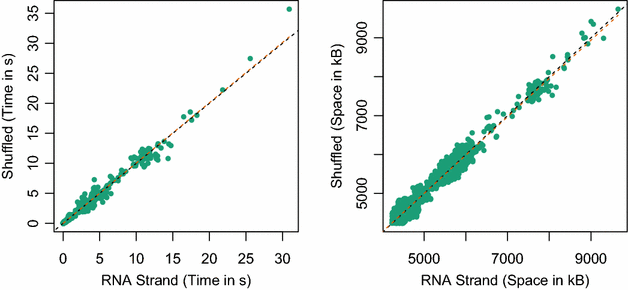
Results: Here, we revisit the problem of space-efficient free energy minimization. Whereas the space-efficient minimization of the free energy has been sketched before, the reconstruction of the optimum structure has not even been discussed. We show that this reconstruction is not possible in trivial extension of the method for simple energy models. Then, we present the time- and space-efficient sparsified free energy minimization algorithm SparseMFEFold that guarantees MFE structure prediction. In particular, this novel algorithm provides efficient fold reconstruction based on dynamically garbage-collected trace arrows. The complexity of our algorithm depends on two parameters, the number of candidates Z and the number of trace arrows T; both are bounded by [Formula: see text], but are typically much smaller. The time complexity of RNA folding is reduced from [Formula: see text] to [Formula: see text]; the space complexity, from [Formula: see text] to [Formula: see text]. Our empirical results show more than 80 % space savings over RNAfold [Vienna RNA package] on the long RNAs from the RNA STRAND database (≥2500 bases).
Conclusions: The presented technique is intentionally generalizable to complex prediction algorithms; due to their high space demands, algorithms like pseudoknot prediction and RNA-RNA-interaction prediction are expected to profit even stronger than "standard" MFE folding. SparseMFEFold is free software, available at http://www.bioinf.uni-leipzig.de/~will/Software/SparseMFEFold.
Keywords: Pseudoknot-free RNA folding; RNA secondary structure prediction; Space efficient sparsification.
Figures



Similar articles
-
SparseRNAfolD: optimized sparse RNA pseudoknot-free folding with dangle consideration.Algorithms Mol Biol. 2024 Mar 3;19(1):9. doi: 10.1186/s13015-024-00256-4. Algorithms Mol Biol. 2024. PMID: 38433200 Free PMC article.
-
Practicality and time complexity of a sparsified RNA folding algorithm.J Bioinform Comput Biol. 2012 Apr;10(2):1241007. doi: 10.1142/S0219720012410077. J Bioinform Comput Biol. 2012. PMID: 22809342
-
An improved Four-Russians method and sparsified Four-Russians algorithm for RNA folding.Algorithms Mol Biol. 2016 Aug 5;11:22. doi: 10.1186/s13015-016-0081-9. eCollection 2016. Algorithms Mol Biol. 2016. PMID: 27499801 Free PMC article.
-
Energy-directed RNA structure prediction.Methods Mol Biol. 2014;1097:71-84. doi: 10.1007/978-1-62703-709-9_4. Methods Mol Biol. 2014. PMID: 24639155 Review.
-
Prediction of RNA secondary structure by free energy minimization.Curr Opin Struct Biol. 2006 Jun;16(3):270-8. doi: 10.1016/j.sbi.2006.05.010. Epub 2006 May 19. Curr Opin Struct Biol. 2006. PMID: 16713706 Review.
Cited by
-
Genetic Polymorphism of miR-196a-2 is Associated with Bone Mineral Density (BMD).Int J Mol Sci. 2017 Nov 25;18(12):2529. doi: 10.3390/ijms18122529. Int J Mol Sci. 2017. PMID: 29186852 Free PMC article.
-
Evolution and structural variations in chloroplast tRNAs in gymnosperms.BMC Genomics. 2021 Oct 18;22(1):750. doi: 10.1186/s12864-021-08058-3. BMC Genomics. 2021. PMID: 34663228 Free PMC article.
-
SparseRNAfolD: optimized sparse RNA pseudoknot-free folding with dangle consideration.Algorithms Mol Biol. 2024 Mar 3;19(1):9. doi: 10.1186/s13015-024-00256-4. Algorithms Mol Biol. 2024. PMID: 38433200 Free PMC article.
-
Role of 4‑aminobutyrate aminotransferase (ABAT) and the lncRNA co‑expression network in the development of myelodysplastic syndrome.Oncol Rep. 2019 Aug;42(2):509-520. doi: 10.3892/or.2019.7175. Epub 2019 May 29. Oncol Rep. 2019. PMID: 31173260 Free PMC article.
References
-
- Backofen R, Tsur D, Zakov S, Ziv-Ukelson M. Sparse RNA folding: time and space efficient algorithms. J Discret Algorithms. 2011;9(1):12–31. doi: 10.1016/j.jda.2010.09.001. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

