De Novo Centromere Formation and Centromeric Sequence Expansion in Wheat and its Wide Hybrids
- PMID: 27110907
- PMCID: PMC4844185
- DOI: 10.1371/journal.pgen.1005997
De Novo Centromere Formation and Centromeric Sequence Expansion in Wheat and its Wide Hybrids
Abstract
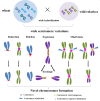
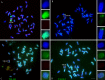
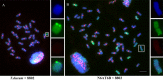
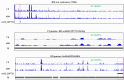
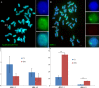
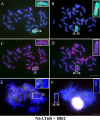
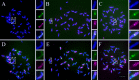
Centromeres typically contain tandem repeat sequences, but centromere function does not necessarily depend on these sequences. We identified functional centromeres with significant quantitative changes in the centromeric retrotransposons of wheat (CRW) contents in wheat aneuploids (Triticum aestivum) and the offspring of wheat wide hybrids. The CRW signals were strongly reduced or essentially lost in some wheat ditelosomic lines and in the addition lines from the wide hybrids. The total loss of the CRW sequences but the presence of CENH3 in these lines suggests that the centromeres were formed de novo. In wheat and its wide hybrids, which carry large complex genomes or no sequenced genome, we performed CENH3-ChIP-dot-blot methods alone or in combination with CENH3-ChIP-seq and identified the ectopic genomic sequences present at the new centromeres. In adcdition, the transcription of the identified DNA sequences was remarkably increased at the new centromere, suggesting that the transcription of the corresponding sequences may be associated with de novo centromere formation. Stable alien chromosomes with two and three regions containing CRW sequences induced by centromere breakage were observed in the wheat-Th. elongatum hybrid derivatives, but only one was a functional centromere. In wheat-rye (Secale cereale) hybrids, the rye centromere-specific sequences spread along the chromosome arms and may have caused centromere expansion. Frequent and significant quantitative alterations in the centromere sequence via chromosomal rearrangement have been systematically described in wheat wide hybridizations, which may affect the retention or loss of the alien chromosomes in the hybrids. Thus, the centromere behavior in wide crosses likely has an important impact on the generation of biodiversity, which ultimately has implications for speciation.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


Similar articles
-
Wheat centromeric retrotransposons: the new ones take a major role in centromeric structure.Plant J. 2013 Mar;73(6):952-65. doi: 10.1111/tpj.12086. Epub 2013 Feb 20. Plant J. 2013. PMID: 23253213
-
Dynamics of the Centromeric Histone CENH3 Structure in Rye-Wheat Amphidiploids (Secalotriticum).Biomed Res Int. 2018 Nov 26;2018:2097845. doi: 10.1155/2018/2097845. eCollection 2018. Biomed Res Int. 2018. PMID: 30598989 Free PMC article.
-
The Parental Centromere Sizes Remain Unaltered in Allopolyploid Wheat-Rye Hybrids.Cytogenet Genome Res. 2024;164(3-4):170-180. doi: 10.1159/000541705. Epub 2024 Oct 1. Cytogenet Genome Res. 2024. PMID: 39353403
-
DNA and proteins of plant centromeres.Curr Opin Plant Biol. 2003 Dec;6(6):554-60. doi: 10.1016/j.pbi.2003.09.007. Curr Opin Plant Biol. 2003. PMID: 14611953 Review.
-
Plant centromeres: genetics, epigenetics and evolution.Mol Biol Rep. 2018 Oct;45(5):1491-1497. doi: 10.1007/s11033-018-4284-7. Epub 2018 Aug 16. Mol Biol Rep. 2018. PMID: 30117088 Review.
Cited by
-
Genetic and Genomic Analyses Reveal Boundaries between Species Closely Related to Cryptococcus Pathogens.mBio. 2019 Jun 11;10(3):e00764-19. doi: 10.1128/mBio.00764-19. mBio. 2019. PMID: 31186317 Free PMC article.
-
Recurrent establishment of de novo centromeres in the pericentromeric region of maize chromosome 3.Chromosome Res. 2017 Oct;25(3-4):299-311. doi: 10.1007/s10577-017-9564-x. Epub 2017 Aug 22. Chromosome Res. 2017. PMID: 28831743
-
Effects of Aegilops longissima chromosome 1Sl on wheat bread-making quality in two types of translocation lines.Theor Appl Genet. 2023 Dec 10;137(1):2. doi: 10.1007/s00122-023-04504-w. Theor Appl Genet. 2023. PMID: 38072878
-
Stable minichromosome and functional neocentromere derived from rye 7R chromosome arm.BMC Plant Biol. 2024 Dec 18;24(1):1185. doi: 10.1186/s12870-024-05918-4. BMC Plant Biol. 2024. PMID: 39695363 Free PMC article.
-
Only the Rye Derived Part of the 1BL/1RS Hybrid Centromere Incorporates CENH3 of Wheat.Front Plant Sci. 2021 Dec 13;12:802222. doi: 10.3389/fpls.2021.802222. eCollection 2021. Front Plant Sci. 2021. PMID: 34966406 Free PMC article.
References
-
- Manuelidis L, Wu JC. Homology between human and simian repeated DNA. Nature. 1978;276: 92–94. - PubMed
-
- Martinez-Zapater JM, Estelle MA, Somerville CR. A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet. 1986;204: 417–423.
-
- Willard HF, Waye JS. Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 1987;3: 192–198.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

