Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity
- PMID: 27111144
- PMCID: PMC4873337
- DOI: 10.1038/ni.3456
Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity
Abstract
The CD4(+) and CD8(+) T cell dichotomy is essential for effective cellular immunity. How individual T cell identity is established remains poorly understood. Here we show that the high-mobility group (HMG) transcription factors Tcf1 and Lef1 are essential for repressing CD4(+) lineage-associated genes including Cd4, Foxp3 and Rorc in CD8(+) T cells. Tcf1- and Lef1-deficient CD8(+) T cells exhibit histone hyperacetylation, which can be ascribed to intrinsic histone deacetylase (HDAC) activity in Tcf1 and Lef1. Mutation of five conserved amino acids in the Tcf1 HDAC domain diminishes HDAC activity and the ability to suppress CD4(+) lineage genes in CD8(+) T cells. These findings reveal that sequence-specific transcription factors can utilize intrinsic HDAC activity to guard cell identity by repressing lineage-inappropriate genes.
Figures
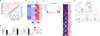
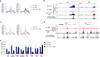

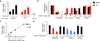
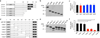


Comment in
-
Tcf1 and Lef1 pack their own HDAC.Nat Immunol. 2016 May 19;17(6):615-6. doi: 10.1038/ni.3469. Nat Immunol. 2016. PMID: 27196513 Free PMC article.
References
-
- Fisher AG. Cellular identity and lineage choice. Nature reviews Immunology. 2002;2(12):977–982. - PubMed
-
- Smale ST. The establishment and maintenance of lymphocyte identity through gene silencing. Nature immunology. 2003;4(7):607–615. - PubMed
-
- Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33(1):12–24. - PubMed
-
- Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes & development. 1995;9(10):1250–1262. - PubMed
References associated with Methods
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

