Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression
- PMID: 27111843
- PMCID: PMC4939857
- DOI: 10.1038/ncb3344
Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression
Abstract
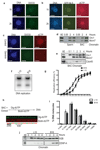
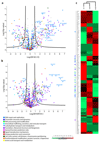
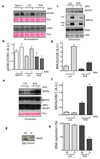
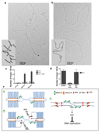
Half of the human genome is made up of repetitive DNA. However, mechanisms underlying replication of chromosome regions containing repetitive DNA are poorly understood. We reconstituted replication of defined human chromosome segments using bacterial artificial chromosomes in Xenopus laevis egg extract. Using this approach we characterized the chromatin assembly and replication dynamics of centromeric alpha-satellite DNA. Proteomic analysis of centromeric chromatin revealed replication-dependent enrichment of a network of DNA repair factors including the MSH2-6 complex, which was required for efficient centromeric DNA replication. However, contrary to expectations, the ATR-dependent checkpoint monitoring DNA replication fork arrest could not be activated on highly repetitive DNA due to the inability of the single-stranded DNA binding protein RPA to accumulate on chromatin. Electron microscopy of centromeric DNA and supercoil mapping revealed the presence of topoisomerase I-dependent DNA loops embedded in a protein matrix enriched for SMC2-4 proteins. This arrangement suppressed ATR signalling by preventing RPA hyper-loading, facilitating replication of centromeric DNA. These findings have important implications for our understanding of repetitive DNA metabolism and centromere organization under normal and stressful conditions.
Figures

Comment in
-
DNA Replication: Looping smoothens repetitive DNA replication.Nat Rev Mol Cell Biol. 2016 Jun;17(6):332. doi: 10.1038/nrm.2016.61. Epub 2016 May 5. Nat Rev Mol Cell Biol. 2016. PMID: 27145719 No abstract available.
-
Replicating repetitive DNA.Nat Cell Biol. 2016 May 27;18(6):593-4. doi: 10.1038/ncb3367. Nat Cell Biol. 2016. PMID: 27230530
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

