Crystal structure of the prefusion surface glycoprotein of the prototypic arenavirus LCMV
- PMID: 27111888
- PMCID: PMC4945123
- DOI: 10.1038/nsmb.3210
Crystal structure of the prefusion surface glycoprotein of the prototypic arenavirus LCMV
Abstract
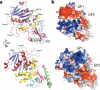
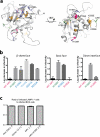
Arenaviruses exist worldwide and can cause hemorrhagic fever and neurologic disease. A single glycoprotein expressed on the viral surface mediates entry into target cells. This glycoprotein, termed GPC, contains a membrane-associated signal peptide, a receptor-binding subunit termed GP1 and a fusion-mediating subunit termed GP2. Although GPC is a critical target of antibodies and vaccines, the structure of the metastable GP1-GP2 prefusion complex has remained elusive for all arenaviruses. Here we describe the crystal structure of the fully glycosylated prefusion GP1-GP2 complex of the prototypic arenavirus LCMV at 3.5 Å. This structure reveals the conformational changes that the arenavirus glycoprotein must undergo to cause fusion and illustrates the fusion regions and potential oligomeric states.
Figures




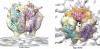
Similar articles
-
Arenavirus stable signal peptide is the keystone subunit for glycoprotein complex organization.mBio. 2014 Oct 28;5(6):e02063. doi: 10.1128/mBio.02063-14. mBio. 2014. PMID: 25352624 Free PMC article.
-
Biological Characterization of Conserved Residues within the Cytoplasmic Tail of the Pichinde Arenaviral Glycoprotein Subunit 2 (GP2).J Virol. 2019 Oct 29;93(22):e01277-19. doi: 10.1128/JVI.01277-19. Print 2019 Nov 15. J Virol. 2019. PMID: 31462569 Free PMC article.
-
Small-Molecule Fusion Inhibitors Bind the pH-Sensing Stable Signal Peptide-GP2 Subunit Interface of the Lassa Virus Envelope Glycoprotein.J Virol. 2016 Jul 11;90(15):6799-807. doi: 10.1128/JVI.00597-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27194767 Free PMC article.
-
The curious case of arenavirus entry, and its inhibition.Viruses. 2012 Jan;4(1):83-101. doi: 10.3390/v4010083. Epub 2012 Jan 13. Viruses. 2012. PMID: 22355453 Free PMC article. Review.
-
Lassa virus glycoprotein: stopping a moving target.Curr Opin Virol. 2018 Aug;31:52-58. doi: 10.1016/j.coviro.2018.05.002. Epub 2018 May 26. Curr Opin Virol. 2018. PMID: 29843991 Free PMC article. Review.
Cited by
-
Identification of Residues in Lassa Virus Glycoprotein Subunit 2 That Are Critical for Protein Function.Pathogens. 2018 Dec 26;8(1):1. doi: 10.3390/pathogens8010001. Pathogens. 2018. PMID: 30587764 Free PMC article.
-
The late endosome-resident lipid bis(monoacylglycero)phosphate is a cofactor for Lassa virus fusion.PLoS Pathog. 2021 Sep 7;17(9):e1009488. doi: 10.1371/journal.ppat.1009488. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34492091 Free PMC article.
-
Structural basis for antibody-mediated neutralization of Lassa virus.Science. 2017 Jun 2;356(6341):923-928. doi: 10.1126/science.aam7260. Science. 2017. PMID: 28572385 Free PMC article.
-
The Role of Receptor Tyrosine Kinases in Lassa Virus Cell Entry.Viruses. 2020 Aug 6;12(8):857. doi: 10.3390/v12080857. Viruses. 2020. PMID: 32781509 Free PMC article.
-
Virus⁻Host Interactions Involved in Lassa Virus Entry and Genome Replication.Pathogens. 2019 Jan 29;8(1):17. doi: 10.3390/pathogens8010017. Pathogens. 2019. PMID: 30699976 Free PMC article. Review.
References
-
- Stephensen CB, et al. Prevalence of serum antibodies against lymphocytic choriomeningitis virus in selected populations from two U.S. cities. J Med Virol. 1992;38:27–31. - PubMed
-
- Childs JE, et al. Human-rodent contact and infection with lymphocytic choriomeningitis and Seoul viruses in an inner-city population. Am J Trop Med Hyg. 1991;44:117–21. - PubMed
-
- Ambrosio AM, Feuillade MR, Gamboa GS, Maiztegui JI. Prevalence of lymphocytic choriomeningitis virus infection in a human population of Argentina. Am J Trop Med Hyg. 1994;50:381–6. - PubMed
-
- Marrie TJ, Saron MF. Seroprevalence of lymphocytic choriomeningitis virus in Nova Scotia. Am J Trop Med Hyg. 1998;58:47–9. - PubMed
-
- Lledo L, Gegundez MI, Saz JV, Bahamontes N, Beltran M. Lymphocytic choriomeningitis virus infection in a province of Spain: analysis of sera from the general population and wild rodents. J Med Virol. 2003;70:273–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

