Targeting the cAMP and Transforming Growth Factor-β Pathway Increases Proliferation to Promote Re-Epithelialization of Human Stem Cell-Derived Retinal Pigment Epithelium
- PMID: 27112176
- PMCID: PMC4922849
- DOI: 10.5966/sctm.2015-0247
Targeting the cAMP and Transforming Growth Factor-β Pathway Increases Proliferation to Promote Re-Epithelialization of Human Stem Cell-Derived Retinal Pigment Epithelium
Abstract
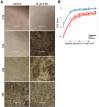
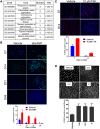
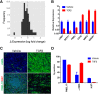
Retinal pigment epithelium (RPE) cell integrity is critical to the maintenance of retinal function. Many retinopathies such as age-related macular degeneration (AMD) are caused by the degeneration or malfunction of the RPE cell layer. Replacement of diseased RPE with healthy, stem cell-derived RPE is a potential therapeutic strategy for treating AMD. Human embryonic stem cells (hESCs) differentiated into RPE progeny have the potential to provide an unlimited supply of cells for transplantation, but challenges around scalability and efficiency of the differentiation process still remain. Using hESC-derived RPE as a cellular model, we sought to understand mechanisms that could be modulated to increase RPE yield after differentiation. We show that RPE epithelialization is a density-dependent process, and cells seeded at low density fail to epithelialize. We demonstrate that activation of the cAMP pathway increases proliferation of dissociated RPE in culture, in part through inhibition of transforming growth factor-β (TGF-β) signaling. This results in enhanced uptake of epithelial identity, even in cultures seeded at low density. In line with these findings, targeted manipulation of the TGF-β pathway with small molecules produces an increase in efficiency of RPE re-epithelialization. Taken together, these data highlight mechanisms that promote epithelial fate acquisition in stem cell-derived RPE. Modulation of these pathways has the potential to favorably impact scalability and clinical translation of hESC-derived RPE as a cell therapy.
Significance: Stem cell-derived retinal pigment epithelium (RPE) is currently being evaluated as a cell-replacement therapy for macular degeneration. This work shows that the process of generating RPE in vitro is regulated by the cAMP and transforming growth factor-β signaling pathway. Modulation of these pathways by small molecules, as identified by phenotypic screening, leads to an increased efficiency of generating RPE cells with a higher yield. This can have a potential impact on manufacturing transplantation-ready cells at large scale and is advantageous for clinical studies using this approach in the future.
Keywords: Proliferation; Retinal pigment epithelium; Stem cells; Transforming growth factor-β; cAMP.
©AlphaMed Press.
Figures




References
-
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. - PubMed
-
- da Cruz L, Chen FK, Ahmado A, et al. RPE transplantation and its role in retinal disease. Prog Retin Eye Res. 2007;26:598–635. - PubMed
-
- Lindekleiv H, Erke MG. Projected prevalence of age-related macular degeneration in Scandinavia 2012-2040. Acta Ophthalmol. 2013;91:307–311. - PubMed
-
- Nazari H, Zhang L, Zhu D, et al. Stem cell based therapies for age-related macular degeneration: The promises and the challenges. Prog Retin Eye Res. 2015;48:1–39. - PubMed
-
- Leach LL, Clegg DO. Concise review: Making stem cells retinal: Methods for deriving retinal pigment epithelium and implications for patients with ocular isease. Stem Cells. 2015;33:2363–2373. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

