Alteration of the serum microbiome composition in cirrhotic patients with ascites
- PMID: 27112233
- PMCID: PMC4845009
- DOI: 10.1038/srep25001
Alteration of the serum microbiome composition in cirrhotic patients with ascites
Abstract
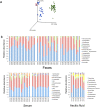
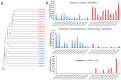
The progression of cirrhosis is associated with alterations in the composition of the gut microbiome. To assess microbial translocation, we compared the serum microbial composition of patients with and without ascites and characterized the ascitic fluid microbiome using 16S rDNA high-throughput sequencing data. A complex and specific microbial community was detected in the serum and ascitic fluid of patients with cirrhosis but barely detectable in the serum of healthy controls. The serum microbiome of patients with ascites presented higher levels of lipopolysaccharide binding protein, a marker of microbial translocation, associated with higher diversity and relative abundance of Clostridiales and an unknown genus belonging to the Cyanobacteria phylum compared to patients without ascites. The composition of the fecal microbiome was also more altered in patients with than without ascites, confirming previous studies on fecal microbiome. We propose that alteration of the serum and fecal microbiome composition be considered indicators of cirrhosis progression.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

