Plant metabolic clusters - from genetics to genomics
- PMID: 27112429
- PMCID: PMC5449196
- DOI: 10.1111/nph.13981
Plant metabolic clusters - from genetics to genomics
Abstract
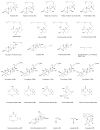
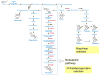
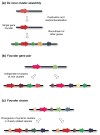
Contents 771 I. 771 II. 772 III. 780 IV. 781 V. 786 786 References 786 SUMMARY: Plant natural products are of great value for agriculture, medicine and a wide range of other industrial applications. The discovery of new plant natural product pathways is currently being revolutionized by two key developments. First, breakthroughs in sequencing technology and reduced cost of sequencing are accelerating the ability to find enzymes and pathways for the biosynthesis of new natural products by identifying the underlying genes. Second, there are now multiple examples in which the genes encoding certain natural product pathways have been found to be grouped together in biosynthetic gene clusters within plant genomes. These advances are now making it possible to develop strategies for systematically mining multiple plant genomes for the discovery of new enzymes, pathways and chemistries. Increased knowledge of the features of plant metabolic gene clusters - architecture, regulation and assembly - will be instrumental in expediting natural product discovery. This review summarizes progress in this area.
Keywords: biosynthetic gene clusters; chromatin; genome mining; natural products; operons.
© 2016 The Authors. New Phytologist © 2016 New Phytologist Trust.
Figures





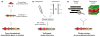
References
-
- Andersen MD, Busk PK, Svendsen I, Møller BL. Cytochromes P450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin. Journal of Biological Chemistry. 2000;275:1966–1975. - PubMed
-
- Arneson PA, Durbin RD. Sensitivity of fungi to alpha-tomatine. Phytopathology. 1968;58:536–537.
-
- Bak S, Nielsen HL, Halkier BA. The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to oxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Molecular Biology. 1998;38:725–734. - PubMed
-
- Balkema-Boomstra AG, Zijlstra S, Verstappen FWA, Inggamer H, Mercke PE, Jongsma MA, Bouwmeester HJ. Role of cucurbitacin C in resistance to spider mite (Tetranychus urticae) in cucumber (Cucumis sativus L.) Journal of Chemical Ecology. 2003;29:225–235. - PubMed
-
- Beaudoin GAW, Facchini PJ. Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta. 2014;240:19–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/00000614/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004561/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M028860/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- U01 GM110699/GM/NIGMS NIH HHS/United States
- BB/L014130/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

