The Treponema denticola FhbB Protein Is a Dominant Early Antigen That Elicits FhbB Variant-Specific Antibodies That Block Factor H Binding and Cleavage by Dentilisin
- PMID: 27113359
- PMCID: PMC4936362
- DOI: 10.1128/IAI.01542-15
The Treponema denticola FhbB Protein Is a Dominant Early Antigen That Elicits FhbB Variant-Specific Antibodies That Block Factor H Binding and Cleavage by Dentilisin
Abstract
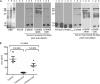
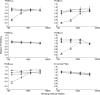
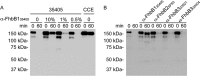
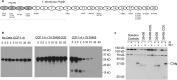
The Treponema denticola FhbB protein contributes to immune evasion by binding factor H (FH). Cleavage of FH by the T. denticola protease, dentilisin, may contribute to the local immune dysregulation that is characteristic of periodontal disease (PD). Although three FhbB phyletic types have been defined (FhbB1, FhbB2, and FhbB3), the in vivo expression patterns and antigenic heterogeneity of FhbB have not been assessed. Here, we demonstrate that FhbB is a dominant early antigen that elicits FhbB type-specific antibody (Ab) responses. Using the murine skin abscess model, we demonstrate that the presence or absence of FhbB or dentilisin significantly influences Ab responses to infection and skin abscess formation. Competitive binding analyses revealed that α-FhbB Ab can compete with FH for binding to T. denticola and block dentilisin-mediated FH cleavage. Lastly, we demonstrate that dentilisin cleavage sites reside within critical functional domains of FH, including the complement regulatory domain formed by CCPs 1 to 4. Analysis of the FH cleavage products revealed that they lack cofactor activity. The data presented here provide insight into the in vivo significance of dentilisin, FhbB and its antigenic diversity, and the potential impact of FH cleavage on the regulation of complement activation.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Plasminogen binding and degradation by Treponema denticola: Identification of the plasminogen binding interface on the FhbB protein.Mol Oral Microbiol. 2018 Jun;33(3):249-256. doi: 10.1111/omi.12221. Epub 2018 Apr 23. Mol Oral Microbiol. 2018. PMID: 29498487
-
Identification of the primary mechanism of complement evasion by the periodontal pathogen, Treponema denticola.Mol Oral Microbiol. 2011 Apr;26(2):140-9. doi: 10.1111/j.2041-1014.2010.00598.x. Epub 2010 Dec 3. Mol Oral Microbiol. 2011. PMID: 21375704 Free PMC article.
-
Analysis of the complement sensitivity of oral treponemes and the potential influence of FH binding, FH cleavage and dentilisin activity on the pathogenesis of periodontal disease.Mol Oral Microbiol. 2014 Oct;29(5):194-207. doi: 10.1111/omi.12054. Epub 2014 Jun 3. Mol Oral Microbiol. 2014. PMID: 24815960 Free PMC article.
-
Approaches to Understanding Mechanisms of Dentilisin Protease Complex Expression in Treponema denticola.Front Cell Infect Microbiol. 2021 May 18;11:668287. doi: 10.3389/fcimb.2021.668287. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34084756 Free PMC article. Review.
-
Creating functional sophistication from simple protein building blocks, exemplified by factor H and the regulators of complement activation.Biochem Soc Trans. 2015 Oct;43(5):812-8. doi: 10.1042/BST20150074. Biochem Soc Trans. 2015. PMID: 26517887 Review.
Cited by
-
The Leptospiral General Secretory Protein D (GspD), a secretin, elicits complement-independent bactericidal antibody against diverse Leptospira species and serovars.Vaccine X. 2021 Feb 23;7:100089. doi: 10.1016/j.jvacx.2021.100089. eCollection 2021 Apr. Vaccine X. 2021. PMID: 33733085 Free PMC article.
-
The leptospiral OmpA-like protein (Loa22) is a surface-exposed antigen that elicits bactericidal antibody against heterologous Leptospira.Vaccine X. 2023 Sep 1;15:100382. doi: 10.1016/j.jvacx.2023.100382. eCollection 2023 Dec. Vaccine X. 2023. PMID: 37727366 Free PMC article.
-
Analysis of the antigenic determinants of the OspC protein of the Lyme disease spirochetes: Evidence that the C10 motif is not immunodominant or required to elicit bactericidal antibody responses.Vaccine. 2019 Apr 17;37(17):2401-2407. doi: 10.1016/j.vaccine.2019.02.007. Epub 2019 Mar 25. Vaccine. 2019. PMID: 30922701 Free PMC article.
-
The major outer sheath protein forms distinct conformers and multimeric complexes in the outer membrane and periplasm of Treponema denticola.Sci Rep. 2017 Oct 16;7(1):13260. doi: 10.1038/s41598-017-13550-6. Sci Rep. 2017. PMID: 29038532 Free PMC article.
-
The Borrelia burgdorferi Adenylate Cyclase, CyaB, Is Important for Virulence Factor Production and Mammalian Infection.Front Microbiol. 2021 May 25;12:676192. doi: 10.3389/fmicb.2021.676192. eCollection 2021. Front Microbiol. 2021. PMID: 34113333 Free PMC article.
References
-
- Miller DP, Bell JK, McDowell JV, Conrad DH, Burgner JW, Heroux A, Marconi RT. 2012. Structure of factor H binding protein B (FhbB) of the periopathogen, Treponema denticola: insights into the progression of periodontal disease. J Biol Chem 287:12715–12722. doi:10.1074/jbc.M112.339721. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

