Folding and Stabilization of Native-Sequence-Reversed Proteins
- PMID: 27113844
- PMCID: PMC4844985
- DOI: 10.1038/srep25138
Folding and Stabilization of Native-Sequence-Reversed Proteins
Abstract
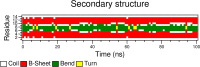
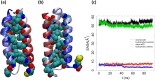
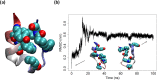
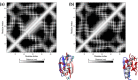
Though the problem of sequence-reversed protein folding is largely unexplored, one might speculate that reversed native protein sequences should be significantly more foldable than purely random heteropolymer sequences. In this article, we investigate how the reverse-sequences of native proteins might fold by examining a series of small proteins of increasing structural complexity (α-helix, β-hairpin, α-helix bundle, and α/β-protein). Employing a tandem protein structure prediction algorithmic and molecular dynamics simulation approach, we find that the ability of reverse sequences to adopt native-like folds is strongly influenced by protein size and the flexibility of the native hydrophobic core. For β-hairpins with reverse-sequences that fail to fold, we employ a simple mutational strategy for guiding stable hairpin formation that involves the insertion of amino acids into the β-turn region. This systematic look at reverse sequence duality sheds new light on the problem of protein sequence-structure mapping and may serve to inspire new protein design and protein structure prediction protocols.
Figures

Similar articles
-
Ultrafast folding and molecular dynamics of a linear hydrophobic β-hairpin.J Biomol Struct Dyn. 2013 Dec;31(12):1404-10. doi: 10.1080/07391102.2012.738612. Epub 2012 Nov 12. J Biomol Struct Dyn. 2013. PMID: 23145986
-
Folding mechanisms of proteins with high sequence identity but different folds.Biochemistry. 2007 Feb 13;46(6):1545-56. doi: 10.1021/bi061904l. Biochemistry. 2007. PMID: 17279619
-
Structure change of β-hairpin induced by turn optimization: an enhanced sampling molecular dynamics simulation study.J Chem Phys. 2011 Dec 21;135(23):235104. doi: 10.1063/1.3668288. J Chem Phys. 2011. PMID: 22191904
-
Understanding the mechanism of beta-sheet folding from a chemical and biological perspective.Biopolymers. 2008;90(6):751-8. doi: 10.1002/bip.21101. Biopolymers. 2008. PMID: 18844292 Review.
-
Using the folding landscapes of proteins to understand protein function.Curr Opin Struct Biol. 2016 Feb;36:67-74. doi: 10.1016/j.sbi.2016.01.001. Epub 2016 Jan 24. Curr Opin Struct Biol. 2016. PMID: 26812092 Review.
Cited by
-
Sequence Reversal Prevents Chain Collapse and Yields Heat-Sensitive Intrinsic Disorder.Biophys J. 2018 Jul 17;115(2):328-340. doi: 10.1016/j.bpj.2018.06.006. Biophys J. 2018. PMID: 30021108 Free PMC article.
-
Reversion of MRAP2 Protein Sequence Generates a Functional Novel Pharmacological Modulator for MC4R Signaling.Biology (Basel). 2022 Jun 7;11(6):874. doi: 10.3390/biology11060874. Biology (Basel). 2022. PMID: 35741395 Free PMC article.
-
Probing Protein Folding with Sequence-Reversed α-Helical Bundles.Int J Mol Sci. 2021 Feb 16;22(4):1955. doi: 10.3390/ijms22041955. Int J Mol Sci. 2021. PMID: 33669383 Free PMC article.
References
-
- Zhou R., Silverman B. D., Royyuru A. K. & Athma P. Spatial profiling of protein hydrophobicity: native vs. decoy structures. Proteins: Structure, Function, and Bioinformatics 52, 561–572 (2003). - PubMed
-
- Kwiecińska J. & Cieplak M. Chirality and protein folding. Journal of Physics: Condensed Matter 17, S1565 (2005).
-
- Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature 437, 640–647 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

