Leukemia-Associated Nup214 Fusion Proteins Disturb the XPO1-Mediated Nuclear-Cytoplasmic Transport Pathway and Thereby the NF-κB Signaling Pathway
- PMID: 27114368
- PMCID: PMC4911740
- DOI: 10.1128/MCB.00158-16
Leukemia-Associated Nup214 Fusion Proteins Disturb the XPO1-Mediated Nuclear-Cytoplasmic Transport Pathway and Thereby the NF-κB Signaling Pathway
Abstract
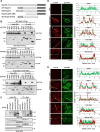
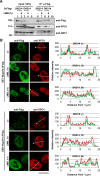
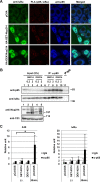
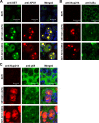
Nuclear-cytoplasmic transport through nuclear pore complexes is mediated by nuclear transport receptors. Previous reports have suggested that aberrant nuclear-cytoplasmic transport due to mutations or overexpression of nuclear pore complexes and nuclear transport receptors is closely linked to diseases. Nup214, a component of nuclear pore complexes, has been found as chimeric fusion proteins in leukemia. Among various Nup214 fusion proteins, SET-Nup214 and DEK-Nup214 have been shown to be engaged in tumorigenesis, but their oncogenic mechanisms remain unclear. In this study, we examined the functions of the Nup214 fusion proteins by focusing on their effects on nuclear-cytoplasmic transport. We found that SET-Nup214 and DEK-Nup214 interact with exportin-1 (XPO1)/CRM1 and nuclear RNA export factor 1 (NXF1)/TAP, which mediate leucine-rich nuclear export signal (NES)-dependent protein export and mRNA export, respectively. SET-Nup214 and DEK-Nup214 decreased the XPO1-mediated nuclear export of NES proteins such as cyclin B and proteins involved in the NF-κB signaling pathway by tethering XPO1 onto nuclear dots where Nup214 fusion proteins are localized. We also demonstrated that SET-Nup214 and DEK-Nup214 expression inhibited NF-κB-mediated transcription by abnormal tethering of the complex containing p65 and its inhibitor, IκB, in the nucleus. These results suggest that SET-Nup214 and DEK-Nup214 perturb the regulation of gene expression through alteration of the nuclear-cytoplasmic transport system.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
