A Novel Quantitative Mass Spectrometry Platform for Determining Protein O-GlcNAcylation Dynamics
- PMID: 27114449
- PMCID: PMC4937517
- DOI: 10.1074/mcp.O115.049627
A Novel Quantitative Mass Spectrometry Platform for Determining Protein O-GlcNAcylation Dynamics
Abstract
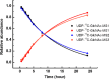
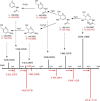
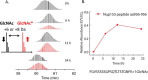
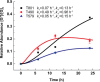
Over the past decades, protein O-GlcNAcylation has been found to play a fundamental role in cell cycle control, metabolism, transcriptional regulation, and cellular signaling. Nevertheless, quantitative approaches to determine in vivo GlcNAc dynamics at a large-scale are still not readily available. Here, we have developed an approach to isotopically label O-GlcNAc modifications on proteins by producing (13)C-labeled UDP-GlcNAc from (13)C6-glucose via the hexosamine biosynthetic pathway. This metabolic labeling was combined with quantitative mass spectrometry-based proteomics to determine protein O-GlcNAcylation turnover rates. First, an efficient enrichment method for O-GlcNAc peptides was developed with the use of phenylboronic acid solid-phase extraction and anhydrous DMSO. The near stoichiometry reaction between the diol of GlcNAc and boronic acid dramatically improved the enrichment efficiency. Additionally, our kinetic model for turnover rates integrates both metabolomic and proteomic data, which increase the accuracy of the turnover rate estimation. Other advantages of this metabolic labeling method include in vivo application, direct labeling of the O-GlcNAc sites and higher confidence for site identification. Concentrating only on nuclear localized GlcNAc modified proteins, we are able to identify 105 O-GlcNAc peptides on 42 proteins and determine turnover rates of 20 O-GlcNAc peptides from 14 proteins extracted from HeLa nuclei. In general, we found O-GlcNAcylation turnover rates are slower than those published for phosphorylation or acetylation. Nevertheless, the rates widely varied depending on both the protein and the residue modified. We believe this methodology can be broadly applied to reveal turnovers/dynamics of protein O-GlcNAcylation from different biological states and will provide more information on the significance of O-GlcNAcylation, enabling us to study the temporal dynamics of this critical modification for the first time.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
A novel strategy for global mapping of O-GlcNAc proteins and peptides using selective enzymatic deglycosylation, HILIC enrichment and mass spectrometry identification.Talanta. 2017 Jul 1;169:195-202. doi: 10.1016/j.talanta.2017.03.049. Epub 2017 Mar 18. Talanta. 2017. PMID: 28411811
-
Quantitative Profiling of Protein O-GlcNAcylation Sites by an Isotope-Tagged Cleavable Linker.ACS Chem Biol. 2018 Aug 17;13(8):1983-1989. doi: 10.1021/acschembio.8b00414. Epub 2018 Jul 30. ACS Chem Biol. 2018. PMID: 30059200
-
O-GlcNAcylation site mapping by (azide-alkyne) click chemistry and mass spectrometry following intensive fractionation of skeletal muscle cells proteins.J Proteomics. 2018 Aug 30;186:83-97. doi: 10.1016/j.jprot.2018.07.005. Epub 2018 Jul 26. J Proteomics. 2018. PMID: 30016717
-
Mass Spectrometry-Based Chemical and Enzymatic Methods for Global Analysis of Protein Glycosylation.Acc Chem Res. 2018 Aug 21;51(8):1796-1806. doi: 10.1021/acs.accounts.8b00200. Epub 2018 Jul 16. Acc Chem Res. 2018. PMID: 30011186 Free PMC article. Review.
-
Proteomic approaches for site-specific O-GlcNAcylation analysis.Bioanalysis. 2014;6(19):2571-80. doi: 10.4155/bio.14.239. Bioanalysis. 2014. PMID: 25411699 Free PMC article. Review.
Cited by
-
Mass Spectrometry for O-GlcNAcylation.Front Chem. 2021 Dec 6;9:737093. doi: 10.3389/fchem.2021.737093. eCollection 2021. Front Chem. 2021. PMID: 34938717 Free PMC article. Review.
-
Ultradeep O-GlcNAc proteomics reveals widespread O-GlcNAcylation on tyrosine residues of proteins.Proc Natl Acad Sci U S A. 2024 Nov 19;121(47):e2409501121. doi: 10.1073/pnas.2409501121. Epub 2024 Nov 12. Proc Natl Acad Sci U S A. 2024. PMID: 39531497 Free PMC article.
-
Multiomics Analysis of Spatially Distinct Stromal Cells Reveals Tumor-Induced O-Glycosylation of the CDK4-pRB Axis in Fibroblasts at the Invasive Tumor Edge.Cancer Res. 2022 Feb 15;82(4):648-664. doi: 10.1158/0008-5472.CAN-21-1705. Cancer Res. 2022. PMID: 34853070 Free PMC article.
-
Global and site-specific analysis of protein glycosylation in complex biological systems with Mass Spectrometry.Mass Spectrom Rev. 2019 Aug;38(4-5):356-379. doi: 10.1002/mas.21586. Epub 2019 Jan 3. Mass Spectrom Rev. 2019. PMID: 30605224 Free PMC article. Review.
-
Design and Preparation of Novel Nitro-Oxide-Grafted Nanospheres with Enhanced Hydrogen Bonding Interaction for O-GlcNAc Analysis.ACS Appl Mater Interfaces. 2022 Oct 26;14(42):47482-47490. doi: 10.1021/acsami.2c15039. Epub 2022 Oct 14. ACS Appl Mater Interfaces. 2022. PMID: 36240223 Free PMC article.
References
-
- Apweiler R., Hermjakob H., and Sharon N. (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 - PubMed
-
- Hart G. W., Housley M. P., and Slawson C. (2007) Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446,1017–1022 - PubMed
-
- Wells L., Vosseller K., and Hart G. W. (2001) Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science 291, 2376–2378 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

