Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires
- PMID: 27114511
- PMCID: PMC4868480
- DOI: 10.1073/pnas.1525510113
Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires
Abstract
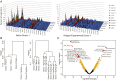
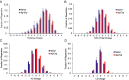
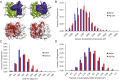
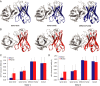
Elucidating how antigen exposure and selection shape the human antibody repertoire is fundamental to our understanding of B-cell immunity. We sequenced the paired heavy- and light-chain variable regions (VH and VL, respectively) from large populations of single B cells combined with computational modeling of antibody structures to evaluate sequence and structural features of human antibody repertoires at unprecedented depth. Analysis of a dataset comprising 55,000 antibody clusters from CD19(+)CD20(+)CD27(-) IgM-naive B cells, >120,000 antibody clusters from CD19(+)CD20(+)CD27(+) antigen-experienced B cells, and >2,000 RosettaAntibody-predicted structural models across three healthy donors led to a number of key findings: (i) VH and VL gene sequences pair in a combinatorial fashion without detectable pairing restrictions at the population level; (ii) certain VH:VL gene pairs were significantly enriched or depleted in the antigen-experienced repertoire relative to the naive repertoire; (iii) antigen selection increased antibody paratope net charge and solvent-accessible surface area; and (iv) public heavy-chain third complementarity-determining region (CDR-H3) antibodies in the antigen-experienced repertoire showed signs of convergent paired light-chain genetic signatures, including shared light-chain third complementarity-determining region (CDR-L3) amino acid sequences and/or Vκ,λ-Jκ,λ genes. The data reported here address several longstanding questions regarding antibody repertoire selection and development and provide a benchmark for future repertoire-scale analyses of antibody responses to vaccination and disease.
Keywords: B cell; antibody; computational modeling; high-throughput sequencing; immunology.
Conflict of interest statement
Conflict of interest statement: G.G., B.J.D., and A.D.E. declare competing financial interests in the form of a patent filed by the University of Texas at Austin.
Figures

References
-
- Murphy K, Travers P, Walport M, Janeway C. Janeway’s Immunobiology. 8th Ed Garland Science; New York: 2012.
-
- Kirkham PM, Schroeder HW., Jr Antibody structure and the evolution of immunoglobulin V gene segments. Semin Immunol. 1994;6(6):347–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

