Paired octamer rings of retinoschisin suggest a junctional model for cell-cell adhesion in the retina
- PMID: 27114531
- PMCID: PMC4868477
- DOI: 10.1073/pnas.1519048113
Paired octamer rings of retinoschisin suggest a junctional model for cell-cell adhesion in the retina
Abstract
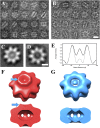
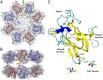
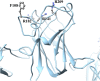
Retinoschisin (RS1) is involved in cell-cell junctions in the retina, but is unique among known cell-adhesion proteins in that it is a soluble secreted protein. Loss-of-function mutations in RS1 lead to early vision impairment in young males, called X-linked retinoschisis. The disease is characterized by separation of inner retinal layers and disruption of synaptic signaling. Using cryo-electron microscopy, we report the structure at 4.1 Å, revealing double octamer rings not observed before. Each subunit is composed of a discoidin domain and a small N-terminal (RS1) domain. The RS1 domains occupy the centers of the rings, but are not required for ring formation and are less clearly defined, suggesting mobility. We determined the structure of the discoidin rings, consistent with known intramolecular and intermolecular disulfides. The interfaces internal to and between rings feature residues implicated in X-linked retinoschisis, indicating the importance of correct assembly. Based on this structure, we propose that RS1 couples neighboring membranes together through octamer-octamer contacts, perhaps modulated by interactions with other membrane components.
Keywords: X-linked retinoschisis; cryo-electron microscopy; discoidin domain; retinoschisin; single particle analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
RS1, a discoidin domain-containing retinal cell adhesion protein associated with X-linked retinoschisis, exists as a novel disulfide-linked octamer.J Biol Chem. 2005 Mar 18;280(11):10721-30. doi: 10.1074/jbc.M413117200. Epub 2005 Jan 11. J Biol Chem. 2005. PMID: 15644328
-
Defective discoidin domain structure, subunit assembly, and endoplasmic reticulum processing of retinoschisin are primary mechanisms responsible for X-linked retinoschisis.J Biol Chem. 2003 Jul 25;278(30):28139-46. doi: 10.1074/jbc.M302464200. Epub 2003 May 13. J Biol Chem. 2003. PMID: 12746437
-
Cog-Wheel Octameric Structure of RS1, the Discoidin Domain Containing Retinal Protein Associated with X-Linked Retinoschisis.PLoS One. 2016 Jan 26;11(1):e0147653. doi: 10.1371/journal.pone.0147653. eCollection 2016. PLoS One. 2016. PMID: 26812435 Free PMC article.
-
Advances in understanding the molecular structure of retinoschisin while questions remain of biological function.Prog Retin Eye Res. 2023 Jul;95:101147. doi: 10.1016/j.preteyeres.2022.101147. Epub 2022 Nov 16. Prog Retin Eye Res. 2023. PMID: 36402656 Free PMC article. Review.
-
Organization and molecular interactions of retinoschisin in photoreceptors.Adv Exp Med Biol. 2008;613:291-7. doi: 10.1007/978-0-387-74904-4_34. Adv Exp Med Biol. 2008. PMID: 18188957 Review. No abstract available.
Cited by
-
Protocols for Processing and Interpreting cryoEM Data Using Bsoft: A Case Study of the Retinal Adhesion Protein, Retinoschisin.Bio Protoc. 2020 Jan 20;10(2):e3491. doi: 10.21769/BioProtoc.3491. eCollection 2020 Jan 20. Bio Protoc. 2020. PMID: 33654723 Free PMC article.
-
Identification of the retinoschisin-binding site on the retinal Na/K-ATPase.PLoS One. 2019 May 2;14(5):e0216320. doi: 10.1371/journal.pone.0216320. eCollection 2019. PLoS One. 2019. PMID: 31048931 Free PMC article.
-
Retinoschisin is linked to retinal Na/K-ATPase signaling and localization.Mol Biol Cell. 2017 Aug 1;28(16):2178-2189. doi: 10.1091/mbc.E17-01-0064. Epub 2017 Jun 14. Mol Biol Cell. 2017. PMID: 28615319 Free PMC article.
-
Long-term rearrangement of retinal structures in a novel mutation of X-linked retinoschisis.Biomed Rep. 2017 Sep;7(3):241-246. doi: 10.3892/br.2017.954. Epub 2017 Jul 27. Biomed Rep. 2017. PMID: 28811895 Free PMC article.
-
Cryo-EM of retinoschisin branched networks suggests an intercellular adhesive scaffold in the retina.J Cell Biol. 2019 Mar 4;218(3):1027-1038. doi: 10.1083/jcb.201806148. Epub 2019 Jan 10. J Cell Biol. 2019. PMID: 30630865 Free PMC article.
References
-
- Haseloff RF, Dithmer S, Winkler L, Wolburg H, Blasig IE. Transmembrane proteins of the tight junctions at the blood-brain barrier: Structural and functional aspects. Semin Cell Dev Biol. 2015;38:16–25. - PubMed
-
- Sosinsky GE, Nicholson BJ. Structural organization of gap junction channels. Biochim Biophys Acta. 2005;1711(2):99–125. - PubMed
-
- Molday RS. Focus on molecules: Retinoschisin (RS1) Exp Eye Res. 2007;84(2):227–228. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

