Homophilic Protocadherin Cell-Cell Interactions Promote Dendrite Complexity
- PMID: 27117416
- PMCID: PMC4856576
- DOI: 10.1016/j.celrep.2016.03.093
Homophilic Protocadherin Cell-Cell Interactions Promote Dendrite Complexity
Abstract
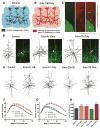
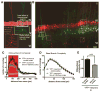
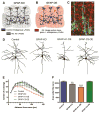
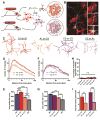
Growth of a properly complex dendrite arbor is a key step in neuronal differentiation and a prerequisite for neural circuit formation. Diverse cell surface molecules, such as the clustered protocadherins (Pcdhs), have long been proposed to regulate circuit formation through specific cell-cell interactions. Here, using transgenic and conditional knockout mice to manipulate γ-Pcdh repertoire in the cerebral cortex, we show that the complexity of a neuron's dendritic arbor is determined by homophilic interactions with other cells. Neurons expressing only one of the 22 γ-Pcdhs can exhibit either exuberant or minimal dendrite complexity, depending only on whether surrounding cells express the same isoform. Furthermore, loss of astrocytic γ-Pcdhs, or disruption of astrocyte-neuron homophilic matching, reduces dendrite complexity cell non-autonomously. Our data indicate that γ-Pcdhs act locally to promote dendrite arborization via homophilic matching, and they confirm that connectivity in vivo depends on molecular interactions between neurons and between neurons and astrocytes.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Balthasar N, Dalgaard LT, Lee CE, Yu J, Funahashi H, Williams T, Ferreira M, Tang V, McGovern RA, Kenny CD, et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell. 2005;123:493–505. - PubMed
-
- Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. - PubMed
-
- Duan X, Krishnaswamy A, De la Huerta I, Sanes JR. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell. 2014;158:793–807. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

