Epigenetic profiles signify cell fate plasticity in unipotent spermatogonial stem and progenitor cells
- PMID: 27117588
- PMCID: PMC4853422
- DOI: 10.1038/ncomms11275
Epigenetic profiles signify cell fate plasticity in unipotent spermatogonial stem and progenitor cells
Abstract
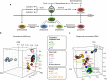
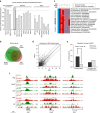
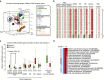
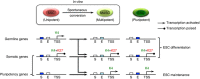
Spermatogonial stem and progenitor cells (SSCs) generate adult male gametes. During in vitro expansion, these unipotent murine cells spontaneously convert to multipotent adult spermatogonial-derived stem cells (MASCs). Here we investigate this conversion process through integrative transcriptomic and epigenomic analyses. We find in SSCs that promoters essential to maintenance and differentiation of embryonic stem cells (ESCs) are enriched with histone H3-lysine4 and -lysine 27 trimethylations. These bivalent modifications are maintained at most somatic promoters after conversion, bestowing MASCs an ESC-like promoter chromatin. At enhancers, the core pluripotency circuitry is activated partially in SSCs and completely in MASCs, concomitant with loss of germ cell-specific gene expression and initiation of embryonic-like programs. Furthermore, SSCs in vitro maintain the epigenomic characteristics of germ cells in vivo. Our observations suggest that SSCs encode innate plasticity through the epigenome and that both conversion of promoter chromatin states and activation of cell type-specific enhancers are prominent features of reprogramming.
Figures



References
-
- Kanatsu-Shinohara M. et al.. Generation of pluripotent stem cells from neonatal mouse testis. Cell 119, 1001–1012 (2004). - PubMed
-
- Takahashi K. & Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). - PubMed
-
- Takahashi K. et al.. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). - PubMed
-
- Pesce M., Wang X., Wolgemuth D. J. & Scholer H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech. Dev. 71, 89–98 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

