Structural insights into ribosome translocation
- PMID: 27117863
- PMCID: PMC4990484
- DOI: 10.1002/wrna.1354
Structural insights into ribosome translocation
Abstract
During protein synthesis, tRNA and mRNA are translocated from the A to P to E sites of the ribosome thus enabling the ribosome to translate one codon of mRNA after the other. Ribosome translocation along mRNA is induced by the universally conserved ribosome GTPase, elongation factor G (EF-G) in bacteria and elongation factor 2 (EF-2) in eukaryotes. Recent structural and single-molecule studies revealed that tRNA and mRNA translocation within the ribosome is accompanied by cyclic forward and reverse rotations between the large and small ribosomal subunits parallel to the plane of the intersubunit interface. In addition, during ribosome translocation, the 'head' domain of small ribosomal subunit undergoes forward- and back-swiveling motions relative to the rest of the small ribosomal subunit around the axis that is orthogonal to the axis of intersubunit rotation. tRNA/mRNA translocation is also coupled to the docking of domain IV of EF-G into the A site of the small ribosomal subunit that converts the thermally driven motions of the ribosome and tRNA into the forward translocation of tRNA/mRNA inside the ribosome. Despite recent and enormous progress made in the understanding of the molecular mechanism of ribosome translocation, the sequence of structural rearrangements of the ribosome, EF-G and tRNA during translocation is still not fully established and awaits further investigation. WIREs RNA 2016, 7:620-636. doi: 10.1002/wrna.1354 For further resources related to this article, please visit the WIREs website.
© 2016 The Authors. WIREs RNA published by Wiley Periodicals, Inc.
Figures

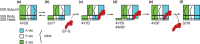




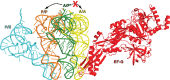
References
FURTHER READING
-
- Frank J. Molecular Machines in Biology: Workshop of the Cell. Cambridge and New York: Cambridge University Press; 2011.
-
- Moore PB. How should we think about the ribosome? Annu Rev Biophys 2012, 41:1–19. - PubMed
-
- Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu Rev Biochem 2013, 82:203–236. - PubMed
References
-
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 2010, 17:555–560. - PubMed
-
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 1997, 385:37–41. - PubMed
-
- Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry 2002, 41:12806–12812. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

