Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency
- PMID: 27118081
- PMCID: PMC4846862
- DOI: 10.1038/srep25222
Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency
Abstract
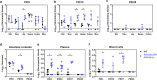
Application of genetically engineered (GE) large animals carrying multi-allelic modifications has been hampered by low efficiency in production and extended gestation period compared to rodents. Here, we rapidly generated RAG2/IL2RG double knockout pigs using direct injection of CRISPR/Cas9 system into developing embryos. RAG2/IL2RG deficient pigs were immunodeficient, characterized by depletion of lymphocytes and either absence of or structurally abnormal immune organs. Pigs were maintained in gnotobiotic facility and evaluated for human norovirus (HuNoV) infection. HuNoV shedding lasted for 16 days in wild type pigs, compared to 27 days (until the end of trials) in RAG2/IL2RG deficient pigs. Additionally, higher HuNoV titers were detected in intestinal tissues and contents and in blood, indicating increased and prolonged HuNoV infection in RAG2/IL2RG deficient pigs and the importance of lymphocytes in HuNoV clearance. These results suggest that GE immunodeficient gnotobiotic pigs serve as a novel model for biomedical research and will facilitate HuNoV studies.
Figures



Similar articles
-
Infectivity of GII.4 human norovirus does not differ between T-B-NK+ severe combined immunodeficiency (SCID) and non-SCID gnotobiotic pigs, implicating the role of NK cells in mediation of human norovirus infection.Virus Res. 2019 Jul 2;267:21-25. doi: 10.1016/j.virusres.2019.05.002. Epub 2019 May 2. Virus Res. 2019. PMID: 31054932 Free PMC article.
-
Enterobacter cloacae inhibits human norovirus infectivity in gnotobiotic pigs.Sci Rep. 2016 Apr 26;6:25017. doi: 10.1038/srep25017. Sci Rep. 2016. PMID: 27113278 Free PMC article.
-
Pathogenesis of Human Norovirus Genogroup II Genotype 4 in Post-Weaning Gnotobiotic Pigs.J Microbiol Biotechnol. 2018 Dec 28;28(12):2133-2140. doi: 10.4014/jmb.1809.09061. J Microbiol Biotechnol. 2018. PMID: 30661347
-
Virus Shedding and Diarrhea: A Review of Human Norovirus Genogroup II Infection in Gnotobiotic Pigs.Viruses. 2024 Sep 7;16(9):1432. doi: 10.3390/v16091432. Viruses. 2024. PMID: 39339908 Free PMC article. Review.
-
Human norovirus cultivation systems and their use in antiviral research.J Virol. 2024 Apr 16;98(4):e0166323. doi: 10.1128/jvi.01663-23. Epub 2024 Mar 12. J Virol. 2024. PMID: 38470106 Free PMC article. Review.
Cited by
-
Generation of RAG2 Knockout Immune-Deficient Miniature Pigs.Animals (Basel). 2024 Sep 6;14(17):2597. doi: 10.3390/ani14172597. Animals (Basel). 2024. PMID: 39272382 Free PMC article.
-
Experimental Methods to Study the Pathogenesis of Human Enteric RNA Viruses.Viruses. 2021 May 25;13(6):975. doi: 10.3390/v13060975. Viruses. 2021. PMID: 34070283 Free PMC article. Review.
-
Fetal bone engraftment reconstitutes the immune system in pigs with severe combined immunodeficiency.Lab Anim (NY). 2024 Oct;53(10):276-286. doi: 10.1038/s41684-024-01439-7. Epub 2024 Sep 17. Lab Anim (NY). 2024. PMID: 39289566 Free PMC article.
-
The history, use, and challenges of therapeutic somatic cell and germline gene editing.Fertil Steril. 2023 Sep;120(3 Pt 1):528-538. doi: 10.1016/j.fertnstert.2023.02.040. Epub 2023 Mar 5. Fertil Steril. 2023. PMID: 36878350 Free PMC article. Review.
-
Human Norovirus: Experimental Models of Infection.Viruses. 2019 Feb 12;11(2):151. doi: 10.3390/v11020151. Viruses. 2019. PMID: 30759780 Free PMC article. Review.
References
-
- Suzuki S. et al. Il2rg gene-targeted severe combined immunodeficiency pigs. Cell Stem Cell 10, 753–8 (2012). - PubMed
-
- Li P. et al. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single-guide RNA and carbohydrate selection. Xenotransplantation 22, 20–31 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

